Step 1: Data selection
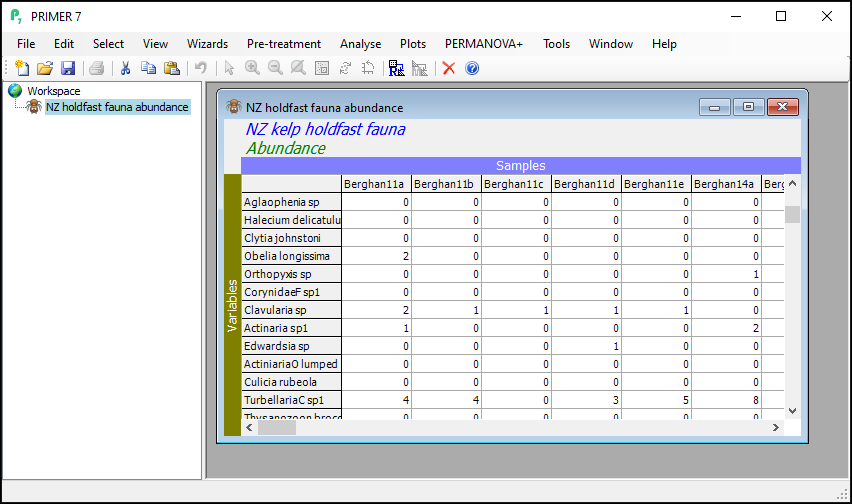
Open up the example data file
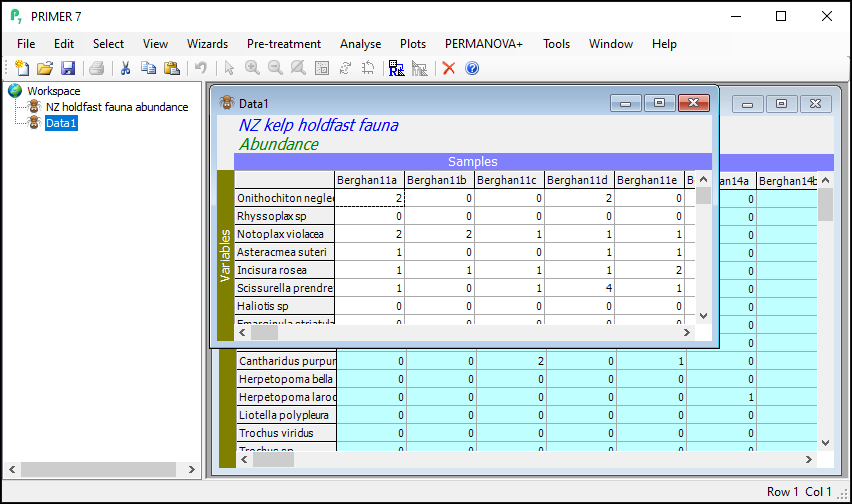
Launch PRIMER, then click File > Open... from the main menu, navigate to the folder named 'NZ holdfast fauna' in the 'Examples v7' directory, and select 'NZ holdfast fauna abundance.pri'. Click Open to display the species matrix.
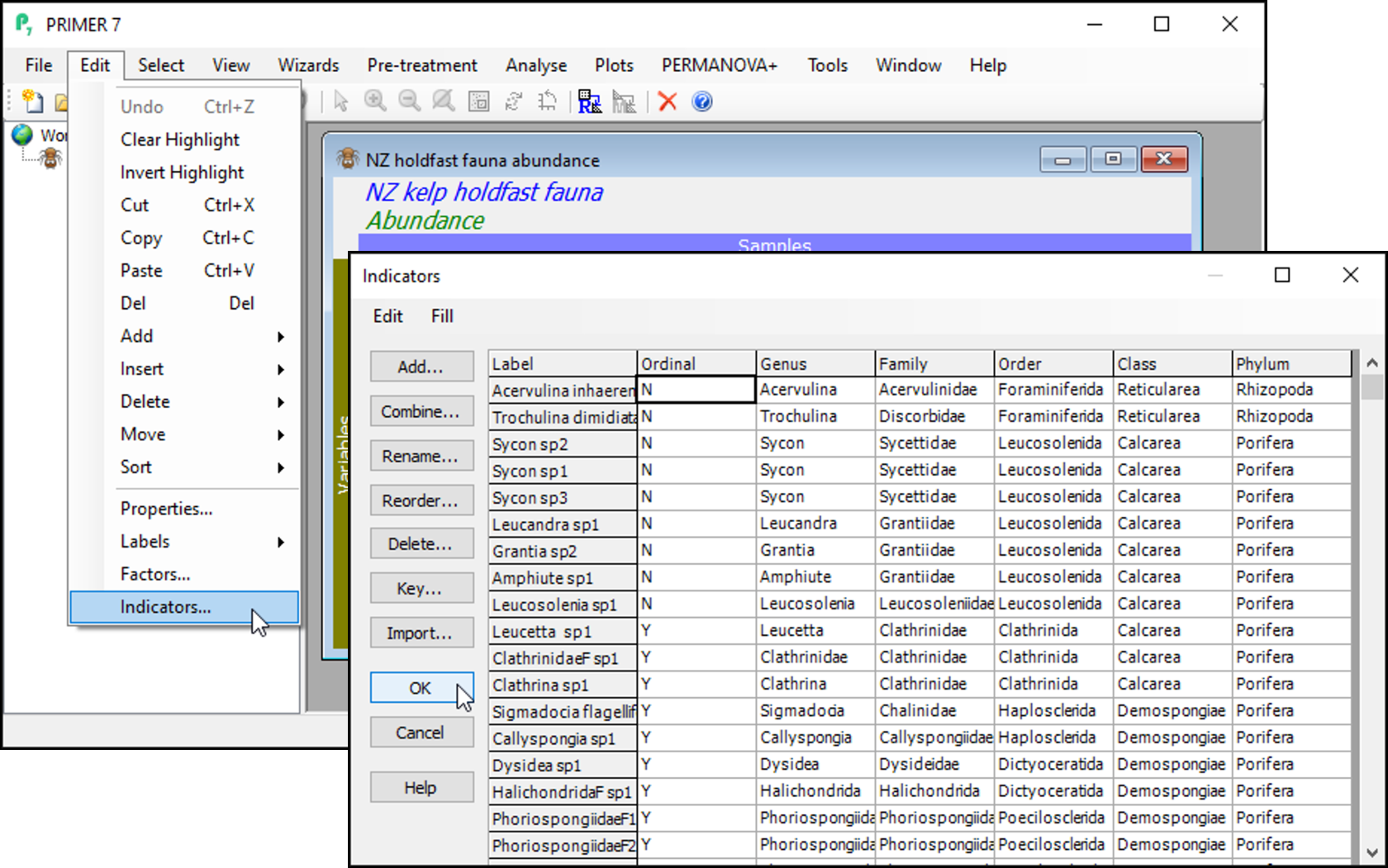
Click on Edit > Indicators... and you will see that the data includes information about whether individual taxa were counted (enumerated) or quantified on an ordinal scale ('Ordinal' = 'N' or 'Y', respectively). Also shown are indicators showing the taxonomic groups in which each species (or taxon) variable belongs, with different levels of the taxonomic hierarchy being provided as different indicators (i.e., 'Genus', 'Family', 'Order', 'Class' and 'Phylum').
Click OK on the 'Indicators' dialog, so the data matrix is the active item in the workspace again.
Select a subset of variables, using an indicator
We wish to select just the mollusc species for the analysis.
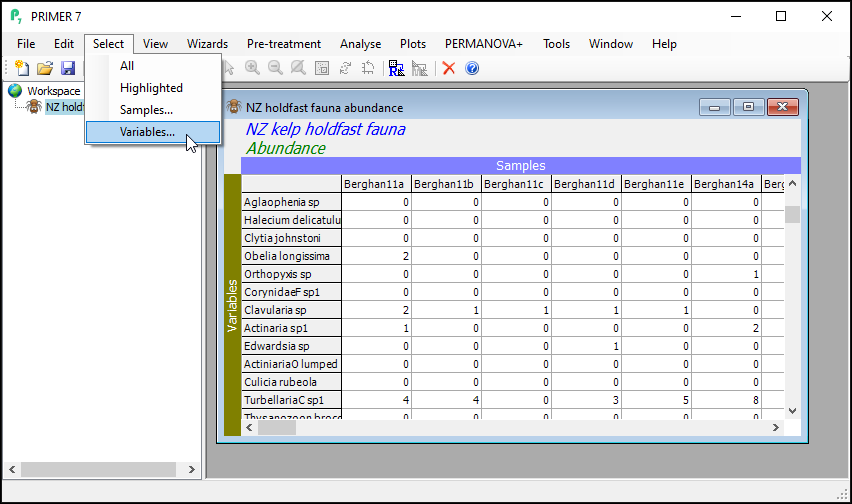
- From the 'NZ holdfast fauna abundance' data sheet, click Select > Variables...
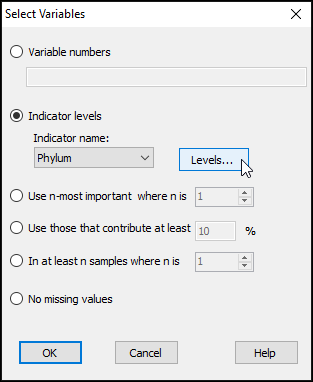
- In the 'Select Variables' dialog, choose ($\bullet$Indicator levels > Indicator name: Phylum) and click Levels....
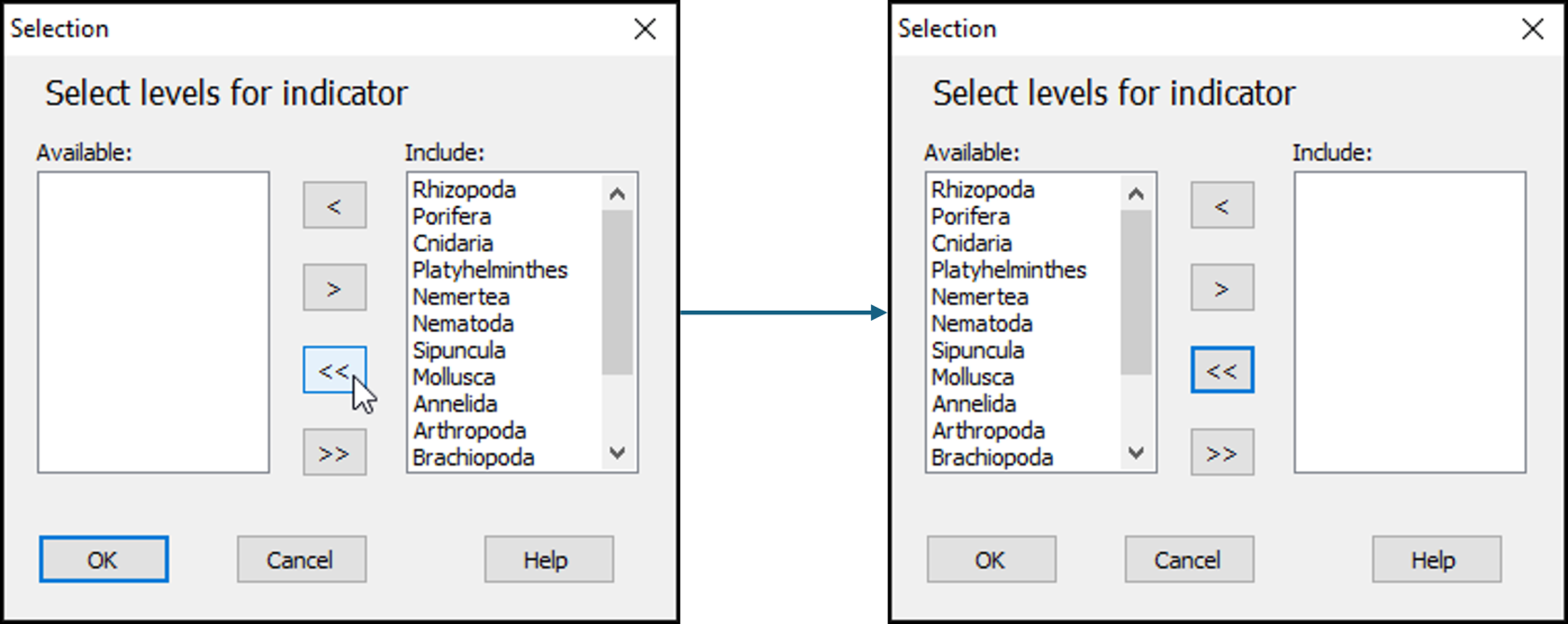
- In the 'Selection' dialog, first move all of the phylum categories from the 'Include:' box (on the right) to the 'Available:' box (on the left) by clicking on the double-left-arrow button:
 .
.
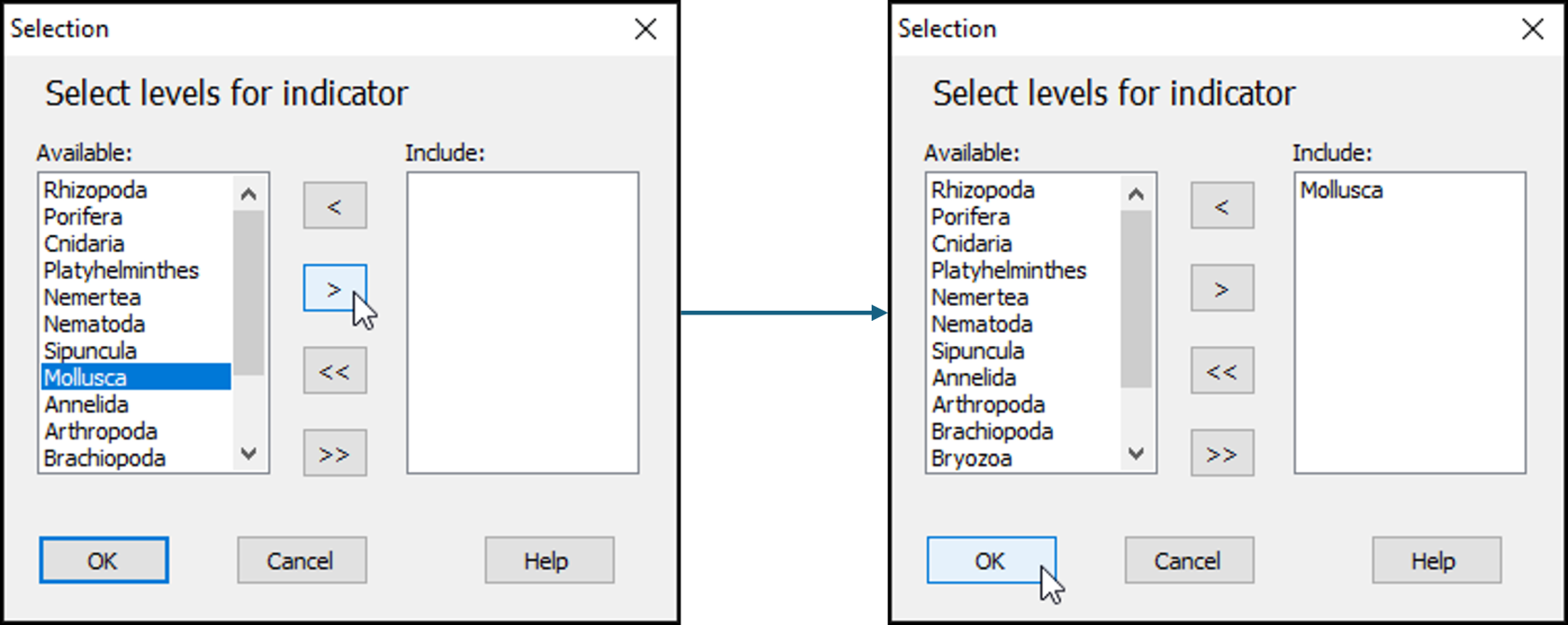
- Next, click on the word 'Mollusca' in the list of 'Available' phylum categories, and click on the single-right-arrow button:
 . This will move it to the 'Include:' box (right-hand side) of the 'Selection' dialog.
. This will move it to the 'Include:' box (right-hand side) of the 'Selection' dialog.
- Click OK on the 'Selection' dialog, then click OK on the 'Select Variables' dialog.
Voila! Whenever you have selected a subset of data (this might be a subset of variables, as done here, or a subset of samples, or both), then the data matrix will have a turquoise background colour to indicate that you have done this, like so:
Any analyses done on a selected subset of data will only be performed on that subset. It is usually a good idea to duplicate and rename a selected subset of data, so as to keep any analysis done on that subset of data clear and separate from the (full) original dataset. Note that subsetting does not affect the original full data matrix of information in any way, which is still always there. You can clear any subset selection (of variables and/or samples) by clicking on Select > All to return to the full data matrix in its entirety. (The turquoise background colour will go away when you do that, and the formerly selected data will yet be highlighted in a purple-ish hue. Clicking on Select > Highlighted can then be used to re-instate the selection from before, if desired.)
Duplicate and rename a selected subset of data
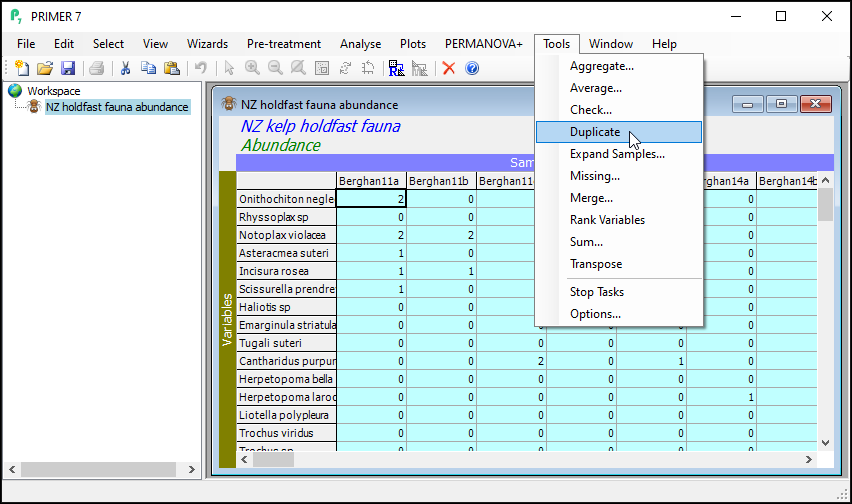
From the subsetted data matrix, click Tools > Duplicate.
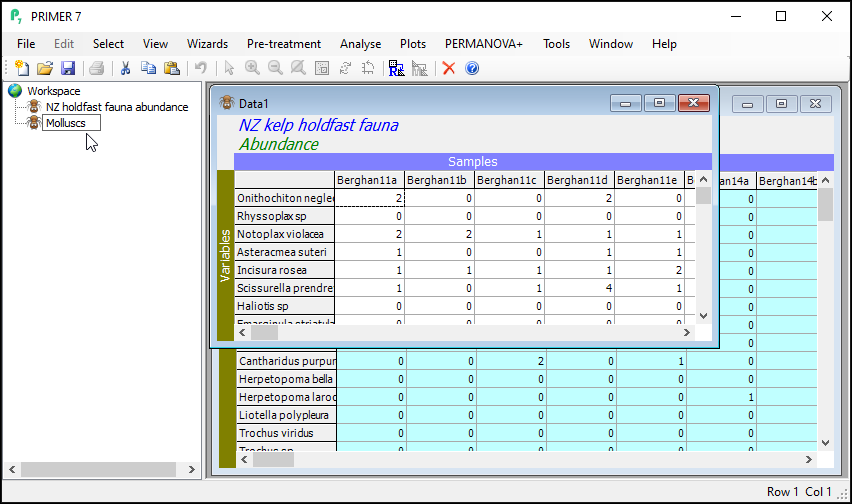
The result will be a new data sheet that contains only the subsetted data (in this case, only the molluscs). It shows up in the Explorer tree window. In the present example, this will (automatically) be called 'Data1'. It is no longer turquoise. The Duplicate routine was performed only on the subsetted (turquoise) data from the original sheet, yeilding this new data sheet, a subset of the original. If you click on Edit > Properties..., you will see that this new sheet now only has 105 variables ('Number of rows:').
Click, hover and click again on the name 'Data1' in the Explorer tree window (or click File > Rename Data or hit the 'F2' key) and type in a new name for the subsetted data sheet: Molluscs.
At this point, you might like to save your workspace. Click File > Save Workspace As... > (Filename: NZ_holdfast_molluscs.pwk).