Step 4: Run PERMANOVA
Once the design file is created, we are ready to go ahead with the PERMANOVA analysis.
- Click on the 'Jaccard' resemblance matrix in the Explorer tree so that it is the active item in the workspace, then click PERMANOVA+ > PERMANOVA...
- Check to see that the 'Design worksheet:' is Design1; this is the design file we created in the previous step that contains the three-way nested design. For the rest, we will keep most of the defaults in the PERMANOVA dialog, but it is wise to increase 'Num. permutations:' from 999 to 9999, as shown below, then click OK.
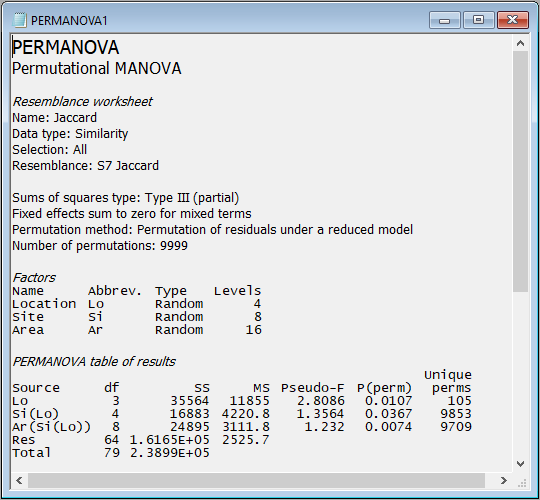
- This produces an output file (called 'PERMANOVA1') in the Explorer tree. It shows:
- the details of the choices you made in the PERMANOVA dialog to run the analysis;
- the details of your experimental design; and
- the PERMANOVA table of results
as follows:
Interpretation
These results show that there is statistically significant variability in the identities of molluscs among holdfasts at each of the three spatial scales in the experimental design: Areas ($F_{8,64}$ = 1.23, $P$ < 0.01), Sites ($F_{4,8}$ = 1.36, $P$ < 0.05) and Locations ($F_{3,4}$ = 2.81, $P$ < 0.05). Note that the p-value for Locations is somewhat limited by the number of unique values of the pseudo-F statistic under permutation that are available here. Specifically, when we permute 2 samples per group (i.e., the 8 Sites) randomly across 4 groups (the Locations), there are just 105 unique values of pseudo-F that can be obtained, so the minimum possible p-value here is 1/105 = 0.0095).