Matrix display
The Matrix display wizard produces a shade plot of a multivariate data matrix, with a useful ordering of its rows and columns that can help to clarify inter-sample and inter-species relationships, as well as gradients in turnover based on a resemblance matrix of choice.
Create a Matrix display
Click on the original data sheet ('Fal nematode abundance', at the top of the Explorer tree window), then click on Wizards > Matrix display....
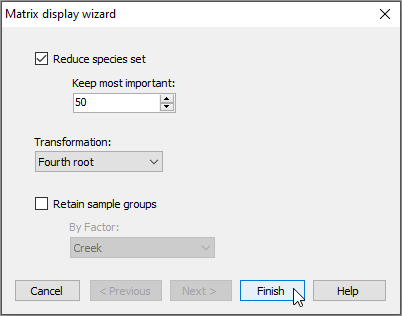
In the 'Matrix display wizard' dialog, leave the defaults, but choose (Transformation: Fourth root), then click Finish.
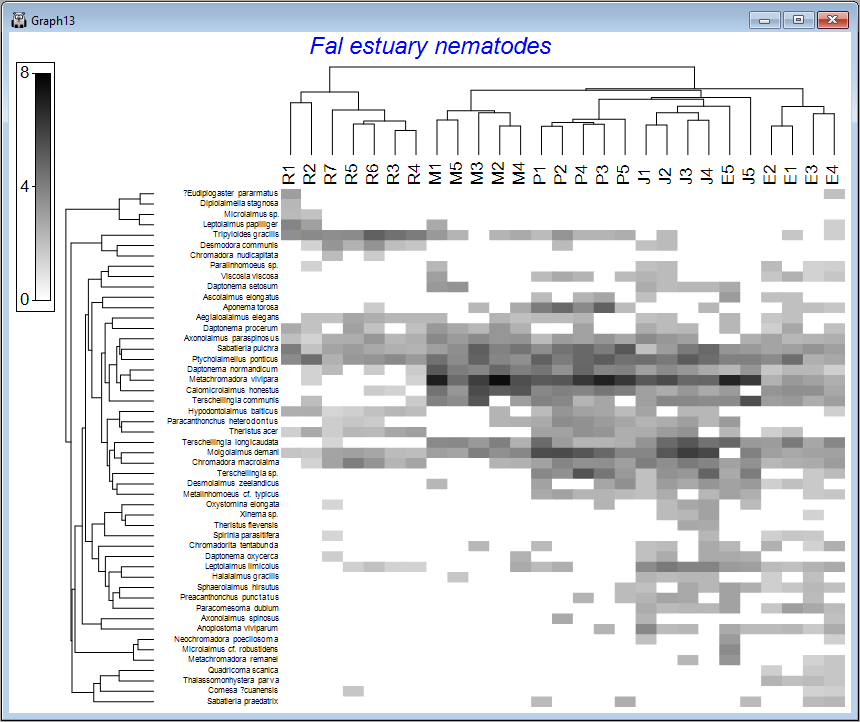
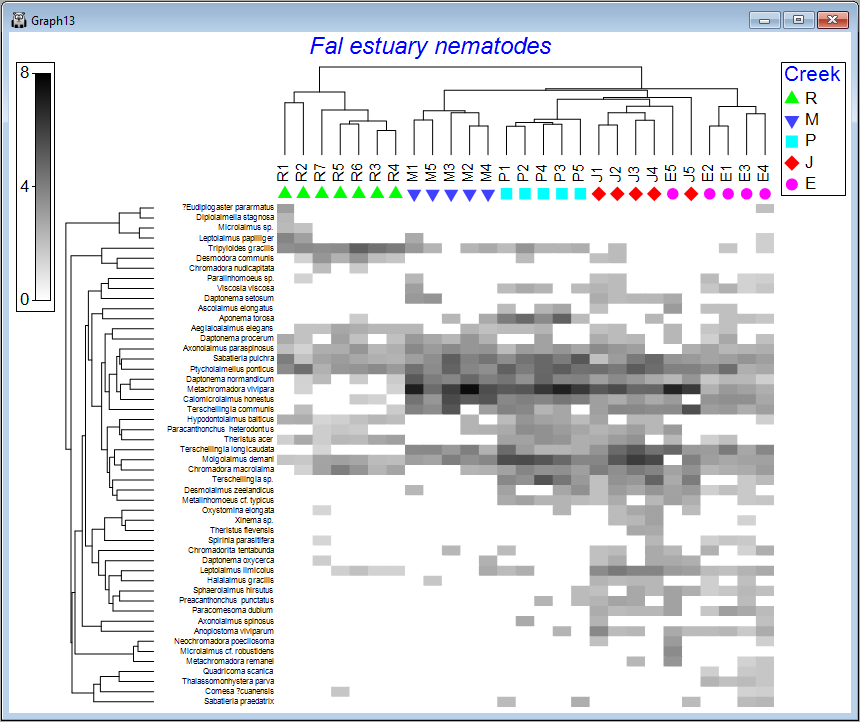
The default graphic ('Graph13')shows a shade plot of the 50 most important species (here, 'most important' is defined based on the percent(%) of the total abundance obtained by a species in any sample). Each value in the data matrix is represented by a shaded rectangle (from white to black, by default), where the degree of shading corresponds to the abundance values (after fourth-root transformation), as shown in the key (in this example, the transformed data values range from 0 to 8).
Here, samples are ordered so as to maximise their concordance with a model of 'seriation' (or turnover), defined on the basis of Bray-Curtis similarities calculated on fourth-root transformed data. They are (furthermore) constrained according to a dendrogram constructed from a hierarchical group-average cluster analysis of those similarities ('Graph12' in the Explorer tree). The species, in turn, are ordered according to a model of seriation based on an Index of Association calculated among species after first standardising the species variables by their total abundance. They are (similarly) further constrained by their own cluster dendrogram ('Graph11' in the Explorer tree).
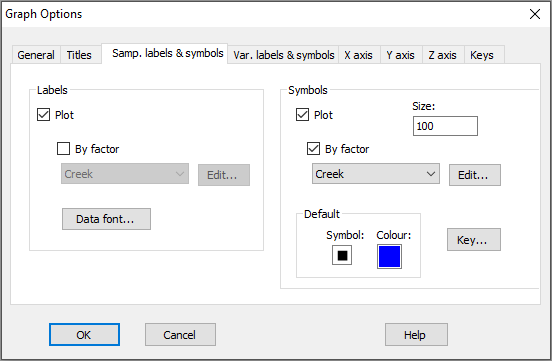
It would perhaps be helpful to see the different creeks as different symbols on this plot. With Graph13 (the shade plot itself) as the active item in the Explorer tree, click Graph > Sample Labels & Symbols, then choose (Labels $\checkmark$Plot) & (Symbols $\checkmark$Plot > $\checkmark$By factor Creek) and click OK.
We can see from the above image that some creeks contain a rather different set of species and/or different abundances of the same species. The ordering of whole creeks shown in the shade plot (obtained using the 'seriation' model) reflects the ordering we saw in the nMDS plot for these creeks as well (see the page 'Step 4. Ordination').
From a matrix display (or shade plot), clicking on Graph > Special reveals a very large number of colours and other graphical options and parameters allowing the user to alter and enhance this graphic for their purposes. These are too numerous to describe in detail here, but they include a host of methods for sorting the rows and/or columns (i.e., by clicking on the Reorder... button in the Graph > Special dialog).