1.4 Sums of squares from a distance matrix
We can now consider the structure of a distance/dissimilarity matrix and how sums of squares for a one-way multivariate ANOVA partitioning would be calculated (Fig. 1.4).
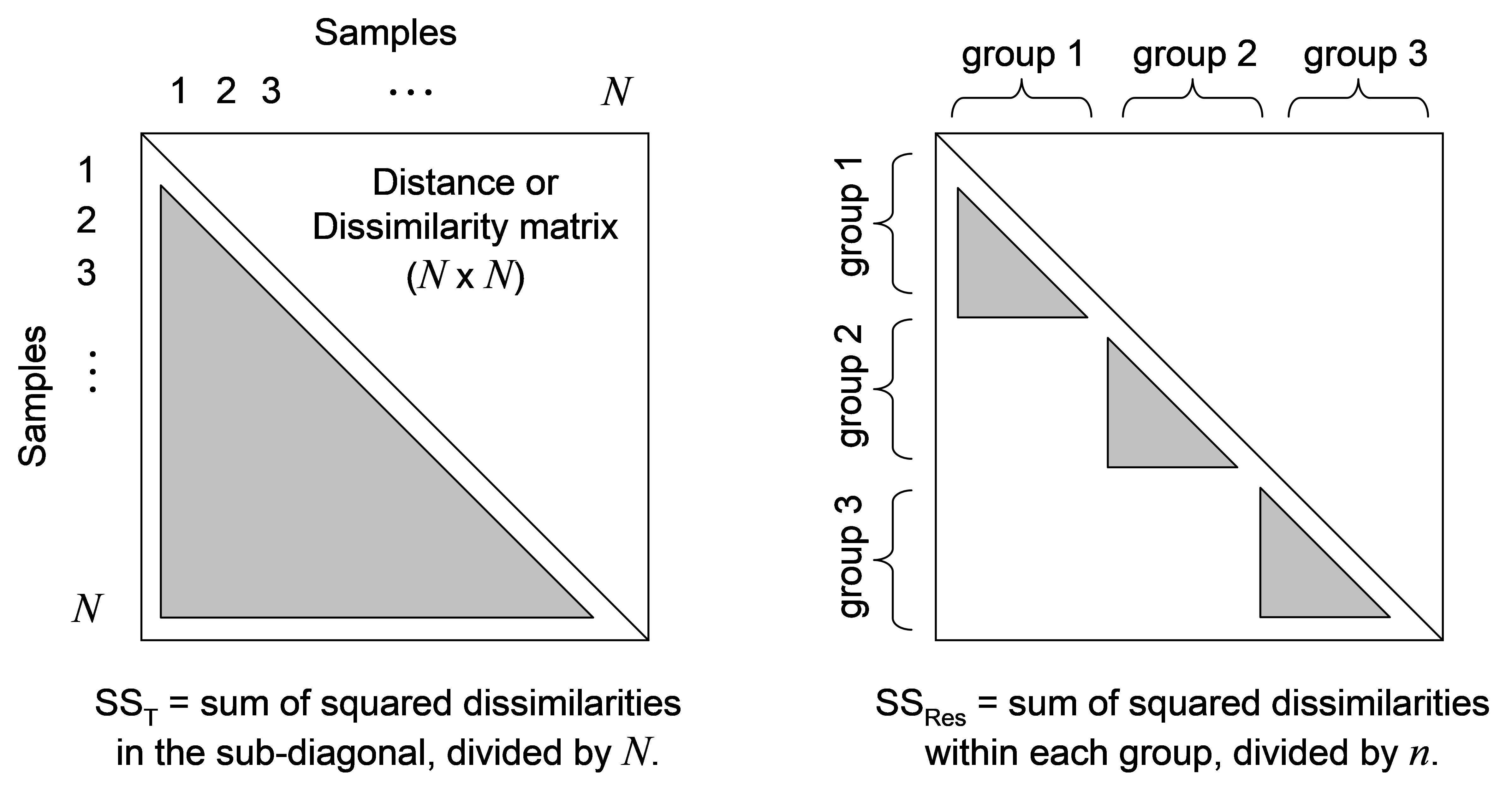 Fig. 1.4. Calculation of sums of squares directly from a distance/dissimilarity matrix.
Fig. 1.4. Calculation of sums of squares directly from a distance/dissimilarity matrix.
If we let $d _ {ij}$ be the dissimilarity (or distance) between sample i and sample j, then the total sum of squares is the sum of the inter-point dissimilarities among all samples, divided by N:
$$ SS _ T = \frac{1}{N} \sum _ {i=1} ^ {N-1} \sum _ {j=i+1} ^ {N} d _ {ij} ^2 \tag{1.1} $$
and the residual (within-group) sum of squares (assuming, for now, a balanced design having an equal sample size of n per group) is:
$$ SS _ {Res} = \frac{1}{n} \sum _ {i=1} ^ {N-1} \sum _ {j=i+1} ^ {N} d _ {ij} ^2 \omega _ {ij} \tag{1.2} $$
where $\omega _ {ij}$ takes the value of 1 if samples i and j are in the same group, otherwise it takes the value of zero. This amounts to adding up the squares of all the dissimilarities between samples that occur within the same group. These quantities are shown schematically in Fig. 1.4. The among-group sum of squares can also be calculated directly or, more simply, as the difference: $SS _A = SS _T - SS _ {Res}$. Partitioning of distance matrices having Euclidean metric properties according to ANOVA experimental designs has been discussed previously by Edgington (1995) , Pillar & Orloci (1996) and Excoffier, Smouse & Quattro (1992) , whereas Gower & Krzanowski (1999) extended this idea to semi-metric dissimilarities having only the properties of symmetry (i.e., $d _{ij} = d _{ji}$) and that $d _ {ij} \ge 0$ and $d _ {ii} = 0$ for all samples.
9 The original simple Mantel test ( Mantel (1967) ) to relate two distance matrices, from which several of the PRIMER routines (RELATE, BIOENV, BEST, BVSTEP and 2-stage MDS) all drew some inspiration, is valid and does have utility in appropriate applications, as pointed out by Legendre, Borcard & Peres-Neto (2005) .
