1.3 Example: Frierfjord macrofauna
The first example is from the IOC/GEEP practical workshop on biological effects of pollutants ( Bayne, Clarke & Gray (1988) ), held at the University of Oslo, August 1986. This attempted to contrast a range of biochemical, cellular, physiological and community analyses, applied to field samples from potentially contaminated and control sites, in a fjordic complex (Frierfjord/Langesundfjord) linked to Oslofjord ({F}, Fig. 1.1). For the benthic macrofaunal component of this study ( Gray, Aschan, Carr et al. (1988) ), four replicate 0.1m2 Day grab samples were taken at each of six sites (A-E and G, Fig 1.1) and, for each sample, organisms retained on a 1.0 mm sieve were identified and counted. Wet weights were determined for each species in each sample, by pooling individuals within species.
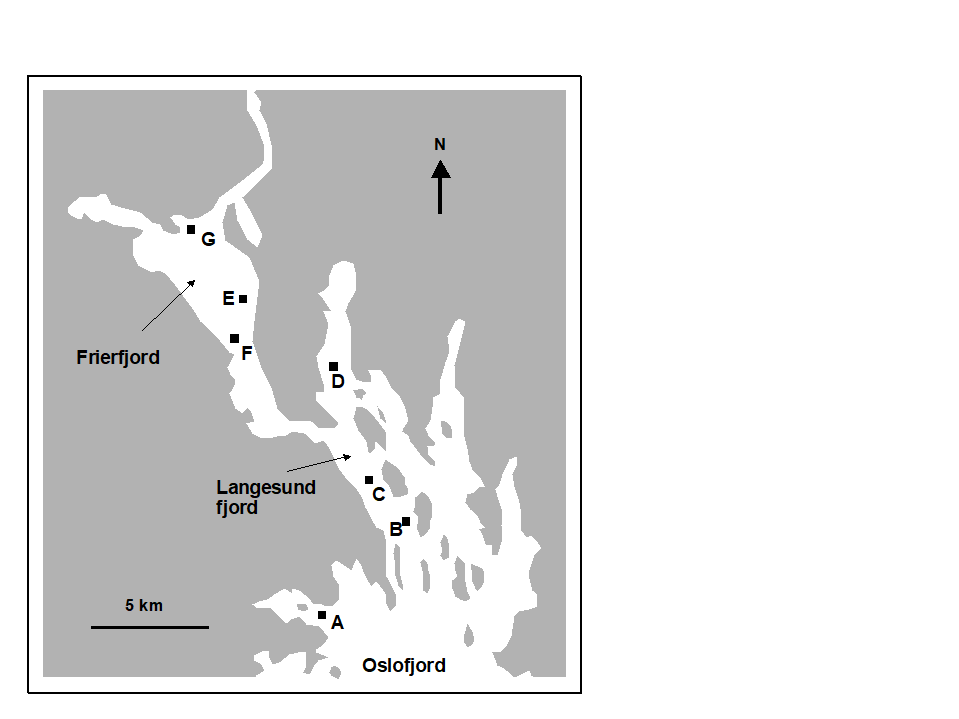 Fig. 1.1. Frierfjord, Norway {F}. Benthic community sampling sites (A-G) for the IOC/GEEP Oslo Workshop; site F omitted for macrobenthos.
Fig. 1.1. Frierfjord, Norway {F}. Benthic community sampling sites (A-G) for the IOC/GEEP Oslo Workshop; site F omitted for macrobenthos.
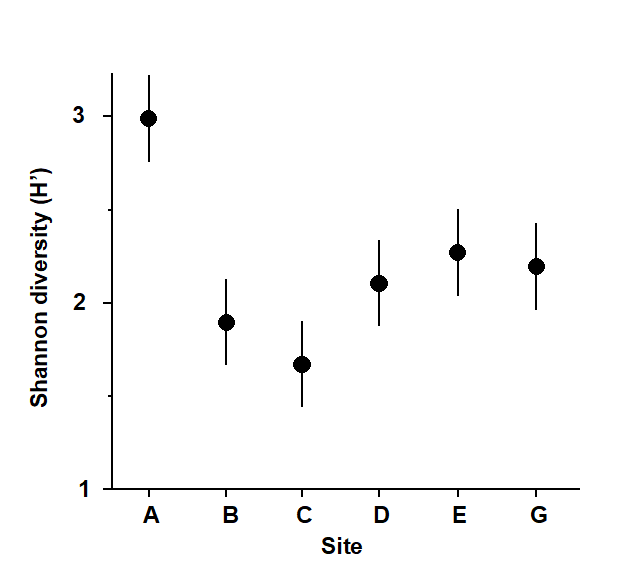
Fig. 1.2. Frierfjord macrofauna {F}. Means and 95% confidence intervals for Shannon diversity (H'), from four replicates at each of six sites (A-E, G).
Part of the resulting data matrix can be seen in Table 1.2: in total there were 110 different taxa categorised from the 24 samples. Such matrices (abundance, A, and/or biomass, B) are the starting point for the biotic analyses of this manual, and this example is typical in respect of the relatively high ratio of species to samples (always >> 1) and the prevalence of zeros. Here, as elsewhere, even an undesirable reduction to the 30 ‘most important’ species (see Chapter 2) leaves more than 50% of the matrix consisting of zeros. Standard multivariate normal analyses (e.g. Mardia, Kent & Bibby (1979) ) of these counts are clearly ruled out; they require both that the number of species (variables) be small in relation to the number of samples, and that the abundance or biomass values are transformable to approximate normality: neither is possible.
Table 1.2. Frierfjord macrofauna {F}. Abundance and biomass matrices (part only) for the 110 species in 24 samples (four replicates at each of six sites A-E, G); abundance in numbers per 0.1m2, biomass in mg per 0.1m2.
| Species | Samples |
|---|
|
A1 | A2 | A3 | A4 | B1 | B2 | B3 | B4 |
|---|---|---|---|---|---|---|---|---|
| Abundance | ||||||||
| Cerianthus lloydi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halicryptus sp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Onchnesoma | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phascolion strombi | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Golfingia sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Holothuroidea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nemertina, indet. | 12 | 6 | 8 | 6 | 40 | 6 | 19 | 7 |
| Polycaeta, indet. | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Amaena trilobata | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Amphicteis gunneri | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| Ampharetidae | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Anaitides groenl. | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Anaitides sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| . . . . | ||||||||
| Biomass | ||||||||
| Cerianthus lloydi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halicryptus sp. | 0 | 0 | 0 | 26 | 0 | 0 | 0 | 0 |
| Onchnesoma | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phascolion strombi | 0 | 0 | 0 | 6 | 0 | 0 | 2 | 0 |
| Golfingia sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Holothuroidea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nemertina, indet. | 1 | 41 | 391 | 1 | 5 | 1 | 2 | 1 |
| Polycaeta, indet. | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Amaena trilobata | 144 | 14 | 234 | 0 | 0 | 0 | 0 | 0 |
| Amphicteis gunneri | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 0 |
| Ampharetidae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anaitides groenl. | 0 | 0 | 0 | 7 | 11 | 0 | 0 | 0 |
| Anaitides sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| . . . . |
As discussed above, one easy route to simplification of this high-dimensional (multi-species) complexity is to reduce each matrix column (sample) to a single univariate description. Fig. 1.2 shows the results of computing the Shannon diversity (H', see Chapter 8) of each sample¶, and plotting for each site the mean diversity and its 95% confidence interval, based on a pooled estimate of variance across all sites from the ANOVA table, Chapter 6. (An analysis of the type outlined in Chapter 9 shows that prior transformation of H' is not required; it already has approximately constant variance across the sites, a necessary prerequisite for standard ANOVA). The most obvious feature of Fig. 1.2 is the relatively higher diversity at the control/reference location, A.
¶ Using the PRIMER DIVERSE routine.