1.7 Multivariate techniques
Table 1.5 summarises some multivariate methods for the four stages, starting with three descriptive tools: hierarchical clustering (agglomerative or divisive), multi-dimensional scaling (MDS, usually non-metric) and principal components analysis (PCA).
Table 1.5. Multivariate techniques. Summary of analyses for the four stages.
The first two of these start explicitly from a triangular matrix of similarity coefficients computed between every pair of samples (e.g. Table 1.6). The coefficient is usually some simple algebraic measure (Chapter 2) of how close the abundance levels are for each species, averaged over all species, and defined such that 100% represents total similarity and 0% complete dissimilarity. There is a range of properties that such a coefficient should possess but still some flexibility in its choice: it is important to realise that the definition of what constitutes similarity of two communities may vary, depending on the biological question under consideration. As with the earlier methods, a multivariate analysis too will attempt to reduce the complexity of the community data by taking a particular ‘view’ of the structure it exhibits. One in which the emphasis is on the pattern of occurrence of rare species will be different than a view in which the emphasis is wholly on the species that are numerically dominant. One convenient way of providing this spectrum of choice, is to restrict attention to a single coefficient†, that of Bray & Curtis (1957) , which has several desirable properties, but allow a choice of prior transformation of the data. A useful transformation continuum (see Chapter 9) ranges through: no transform, square root, fourth root, logarithmic and finally, reduction of the sample information to the recording only of presence or absence for each species.¶ At the former end of the spectrum all attention will be focused on dominant counts, at the latter end on the rarer species.
Table 1.6. Frierfjord macrofauna {F}. Bray-Curtis similarities, after $\sqrt{}\sqrt{}$-transformation of counts, for every pair of replicate samples from sites A, B, C only (four replicates per site).
| A1 | A2 | A3 | A4 | B1 | B2 | B3 | B4 | C1 | C2 | C3 | C4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | – | |||||||||||
| A2 | 61 | – | ||||||||||
| A3 | 69 | 60 | – | |||||||||
| A4 | 65 | 61 | 66 | – | ||||||||
| B1 | 37 | 28 | 37 | 35 | – | |||||||
| B2 | 42 | 34 | 31 | 32 | 55 | – | ||||||
| B3 | 45 | 39 | 39 | 44 | 66 | 66 | – | |||||
| B4 | 37 | 29 | 29 | 37 | 59 | 63 | 60 | – | ||||
| C1 | 35 | 31 | 27 | 25 | 28 | 56 | 40 | 34 | – | |||
| C2 | 40 | 34 | 26 | 29 | 48 | 69 | 62 | 56 | 56 | – | ||
| C3 | 40 | 31 | 37 | 39 | 59 | 61 | 67 | 53 | 40 | 66 | – | |
| C4 | 36 | 28 | 34 | 37 | 65 | 55 | 69 | 55 | 38 | 64 | 74 | – |
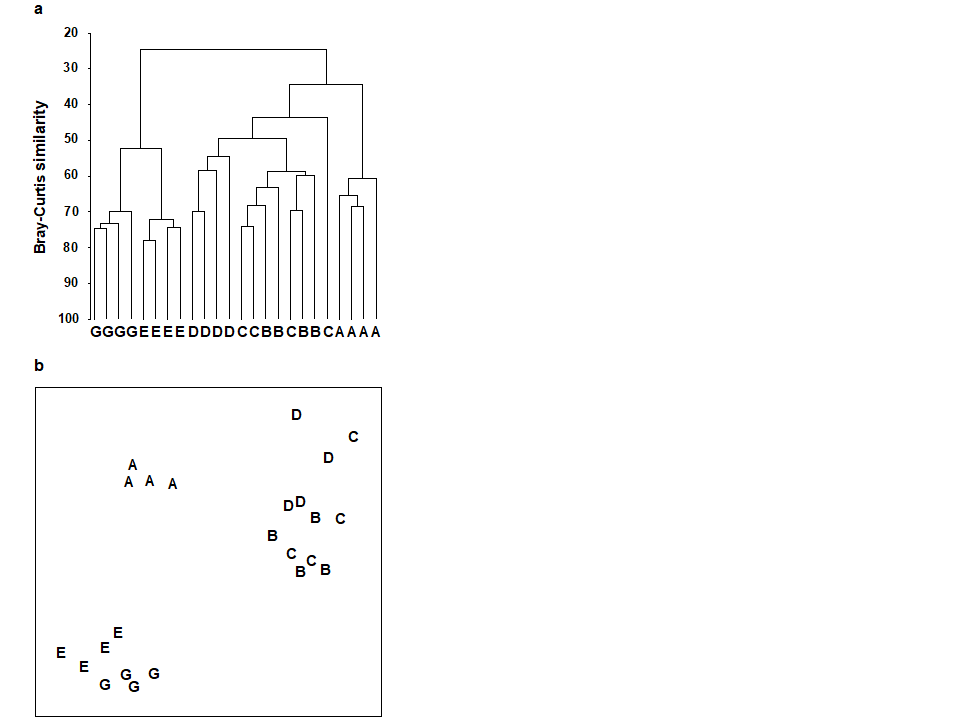
For the clustering technique, representation of the communities for each sample is by a dendrogram (e.g. Fig. 1.7a), linking the samples in hierarchical groups on the basis of some definition of similarity between each cluster (Chapter 3). This is a particularly relevant representation in cases where the samples are expected to divide into well-defined groups, perhaps structured by some clear-cut environmental distinctions. Where, on the other hand, the community pattern is responding to abiotic gradients which are more continuous, then representation by an ordination is usually more appropriate. The method of non-metric MDS (Chapter 5) attempts to place the samples on a ‘map’, usually in two dimensions (e.g. see Fig. 1.7b), in such a way that the rank order of the distances between samples on the map exactly agrees with the rank order of the matching (dis)similarities, taken from the triangular similarity matrix. If successful, and success is measured by a stress coefficient which reflects lack of agreement in the two sets of ranks, the ordination gives a simple and compelling visual representation of ‘closeness’ of the species composition for any two samples.
Fig. 1.7. Frierfjord macrofauna {F}. a) Dendrogram for hierarchical clustering (group-average linking); b) non-metric multi-dimensional scaling (MDS) ordination in two dimensions; both computed for the four replicates from each of the six sites (A–E, G), using the similarity matrix partially shown in Table 1.4 (2-d MDS stress = 0.08)
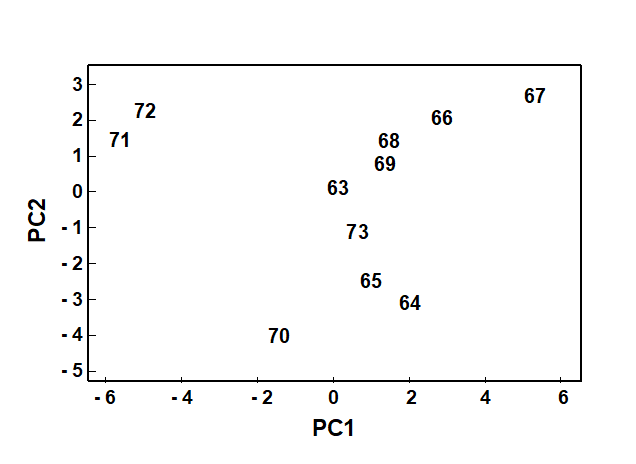
The PCA technique (Chapter 4) takes a different starting position, and makes rather different assumptions about the definition of (dis)similarity of two samples, but again ends up with an ordination plot, often in two or three dimensions (though it could be more), which approximates the continuum of relationships among samples (e.g. Fig. 1.8). In fact, PCA is a rather unsatisfactory procedure for most species-by-samples matrices, for at least two reasons:
a) it defines dissimilarity of samples in an inflexible way (Euclidean distance in the full-dimensional species space, Chapter 4), not well-suited to the rather special nature of species abundance data, with its predominance of zero values;
b) it uses a projection from the higher-dimensional to lower-d space which does not aim to preserve the relative values of these Euclidean distances in the low-d plot, cf MDS, which has that rationale.
Fig. 1.8. Loch Linnhe macrofauna {L}. 2-dimensional principal components analysis (PCA) ordination of the $\sqrt{} \sqrt{}$-transformed abundances from the 11 years 1963–1973 (% of variance explained only 57%, and not an ideal technique for such data).
However, a description of the operation of PCA is included here because it is an historically important technique, the first ordination method to be devised and one which is still commonly encountered§, and because it comes into its own in the analysis of environmental samples. Abiotic variables (e.g. physical or contaminant readings) are usually relatively few in number, continuously scaled, and their distributions can be transformed so that (normalised) Euclidean distances are appropriate ways of describing the inter-relationships among samples. PCA is then a more satisfactory low-dimensional summary (albeit still a projection), and even has an advantage over MDS of providing an interpretation of the plot axes (which are linear in the abiotic variables).
Discriminating sites/conditions from a multivariate analysis requires non-classical hypothesis testing ideas, since it is totally invalid to make the standard assumptions of normality (which in this case would need to be multivariate normality of the sometimes hundreds or even thousands of different species!). Instead, Chapter 6 describes a simple permutation or randomisation test (of the type first developed by Mantel (1967) ), which makes very few assumptions about the data and is therefore widely applicable. In Fig. 1.7b for example, it is clear without further testing that site A has a different community composition across its replicates than the groups (E, G) or (B, C, D). Much less clear is whether there is any statistical evidence of a distinction between the B, C and D sites. A non-parametric test of the null hypothesis of ‘no site differences between B, C and D’ could be constructed by defining a statistic which contrasts among-site and within-site distances, which is then recomputed for all possible permutations of the 12 labels (4 Bs, 4 Cs and 4 Ds) among the 12 locations on the MDS. If these arbitrary site relabellings can generate values of the test statistic which are similar to the value for the real labelling, then there is clearly little evidence that the sites are biologically distinguishable. This idea is formalised and extended to more complex sample designs in Chapter 6. For reasons which are described there it is preferable to compute an ‘among versus within site’ summary statistic directly from the (rank) similarity matrix rather than the distances on the MDS plot. This, and the analogy with ANOVA, suggests the term ANOSIM for the test (Analysis of Similarities, Clarke & Green (1988) ; Clarke (1993) ).‡ It is possible to employ the same test in connection with PCA, using an underlying dissimilarity matrix of Euclidean distances, though when the ordination is of a relatively small number of environmental variables, which can be transformed into approximate multivariate normality, then abiotic differences between sites can use a classical test (MANOVA, e.g. Mardia, Kent & Bibby (1979) ), a generalisation of ANOVA.
Part of the process of discriminating sites, times, treatments etc., where successful, is the ability to identify the species that are principally responsible for these distinctions: it is all too easy to lose sight of the basic data matrix in a welter of sophisticated multivariate analyses of samples.⸸ Similarly, as a result of cluster analyses and associated a posteriori tests for the significance of the groups of sites/times etc obtained (SIMPROF, Chapter 3), one would want to identify the species mainly responsible for distinguishing the clusters from each other. Note the distinction here between a priori groups, identified before examination of the data, for which ANOSIM tests are appropriate (Chapter 6), and a posteriori groups with membership identified as a result of looking at the data, for which ANOSIM is definitely invalid; they need SIMPROF.
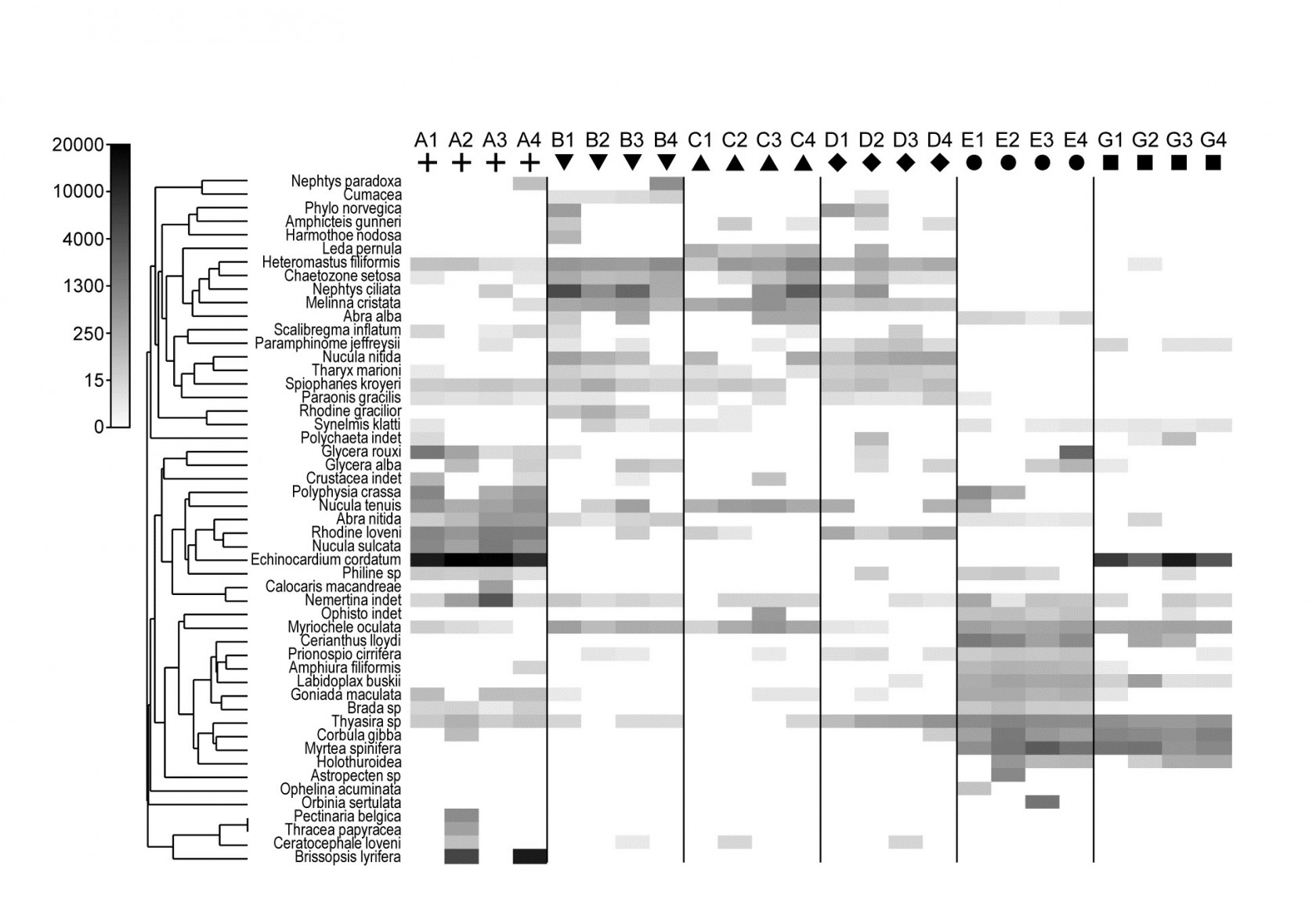
Fig. 1.9. Frierfjord macrofauna {F}. Shade plot of 4th-root transformed species (rows) $\times$ samples (columns) matrix of abundances for the 4 replicate samples at each of 6 sites (Fig. 1.1, Table 1.2). The (linear) grey scale is shown in the key with back-transformed counts.
Species analyses and displays are pursued in Chapter 7, and Fig. 1.9 gives a Shade Plot for the ‘most important’ ~50 species from the 110 recorded from the 24 samples of the Frierfjord macrobenthic abundance data of Table 1.2. (‘Most important’ is here defined as all the species which account for at least 1% of the total abundance in one or more of the samples). The shade plot is a visual representation of the data matrix, after it has been 4th-root transformed, in which white denotes absence and black the largest (transformed) abundance in the data. Importantly, the species axis has been re-ordered in line with a (displayed) cluster analysis of the species, utilising Whittaker’s Index of Association to give the among-species similarities, see Chapters 2 and 7. The pattern of differences between samples from the differing sites is clearly apparent, at least for the three main groups seen in the MDS plot of Fig. 1.7, viz. A, (B-D), (E-G). Such plots are also very useful in visualising the effects of different transformations on the data matrix, prior to similarity computation (see Clarke, Tweedley & Valesini (2014) and Chapter 9). Without transformation, the shade plot would be largely white space with only a handful of species even visible (and thus contributing).
Since ANOSIM indicates statistical significance and pairwise tests give particular site differences (Chapter 6), a ranking of species contributions to the dissimilarity between any specific pair of groups can be obtained from a similarity percentage breakdown (the SIMPER routine, Clarke (1993) ), see Chapter 7.⸙
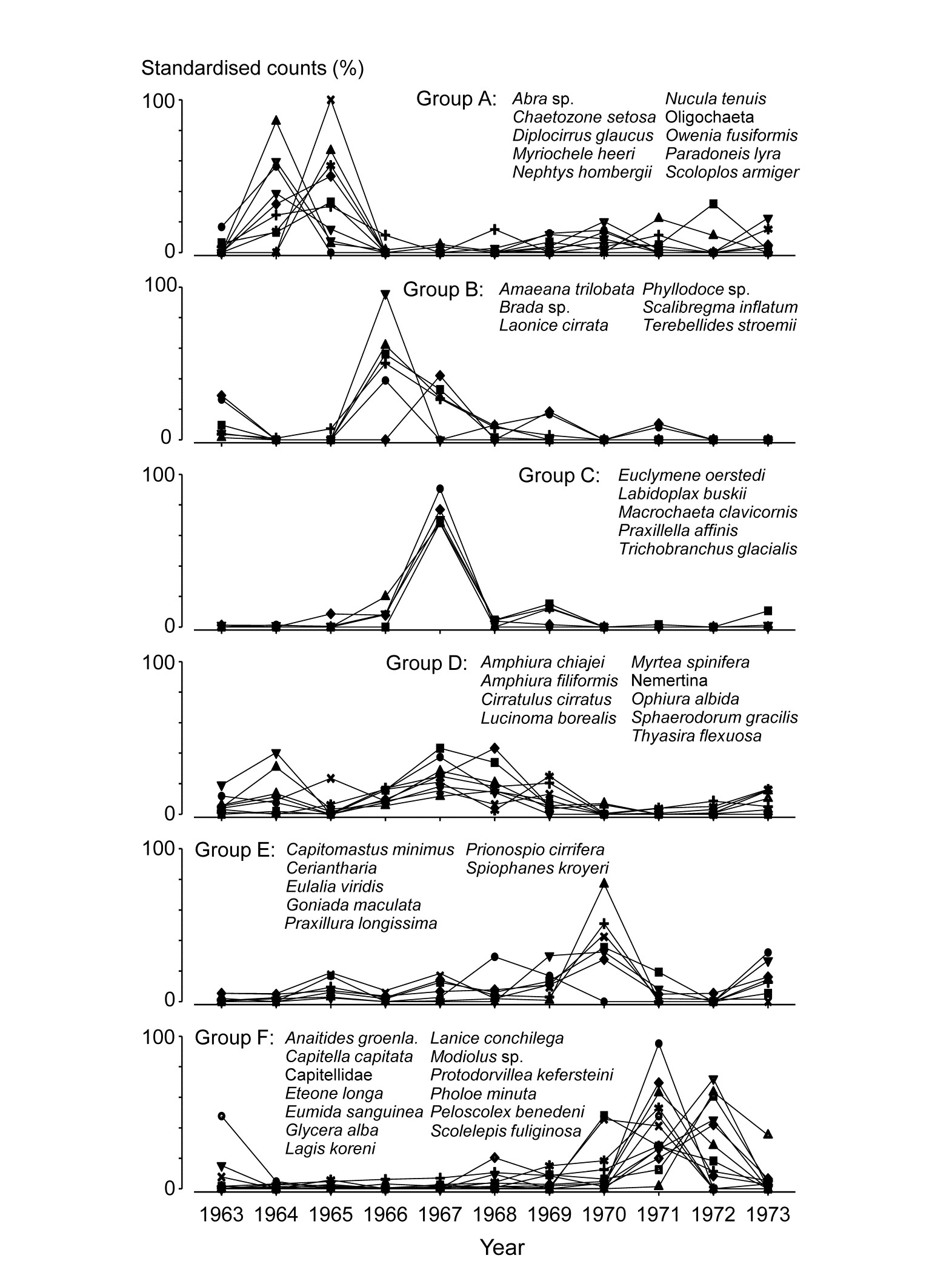
The clustering of species in shade plots such as Fig. 1.9 can be taken one stage further, to determine statistical significance of species groupings (a Type 3 SIMPROF test, see Chapter 7). This identifies groups of species within which the species have statistically indistinguishable patterns of abundance across the set of samples, and between which the patterns do differ significantly. Fig. 1.10 shows simple line plots for the standardised abundance of 51 species (those accounting for > 1% of the total abundance in any one year) over the 11 years of the Loch Linnhe sampling of Table 1.4 and Fig. 1.8. SIMPROF tests give 7 groups of species (one omitted contains just a single species found only in 1973). The standardisation puts each species on an equal footing, with its values summing to 100% across all samples. It can be seen how some species start to disappear, and others arrive, at the initial levels of disturbance, in the mid-years – some of the latter dying out as pollution increases in the later years – with further opportunists (Capitellids etc) flourishing at that point, and then declining with the improvement in conditions in 1973.
Fig. 1.10. Loch Linnhe macrofauna {L}. Line plots of the 11-year time series for the ‘most important’ 51 species (see text), with y axis the standardised counts for each species, i.e. all species add to 100% across years. The 6 species groups (A-F), and a 7th consisting of a single species found in only one year, have internally indistinguishable curves (‘coherent species’) but the sets differ significantly from each other, by SIMPROF tests.
In the determination of stress levels, whilst the multivariate techniques are sensitive (Chapter 14) and well-suited to establishing community differences associated with different sites/times/treatments etc., their species-specific basis would appear to make them unsuitable for drawing general inferences about the pollution status of an isolated group of samples. Even in comparative studies, on the face of it there is not a clear sense of directionality of change when it is established that communities at putatively impacted sites differ from those at control or reference sites in space or time (is the change ‘good’ or ‘bad’?). Nonetheless, there are a few ways in which directionality has been asserted in published studies, whilst retaining a multivariate form of analysis (Chapter 15):
a) a meta-analysis: a combined ordination of data from NE Atlantic shelf waters, at a coarse level of taxonomic discrimination (the effects of taxonomic aggregation are discussed in Chapter 10), suggests a common directional change in the balance of taxa under a variety of types of pollution or disturbance ( Warwick & Clarke (1993a) );
b) a number of studies demonstrate increased multivariate dispersion among replicates under impacted conditions, in comparison to controls ( Warwick & Clarke (1993b) );
c) another feature of disturbance, demonstrated in a spatial coral community study (but with wider applicability to other spatial and temporal patterns), is a loss of smooth seriation along transects of increasing depth, again in comparison to reference data in time and space ( Clarke, Warwick & Brown (1993) ).
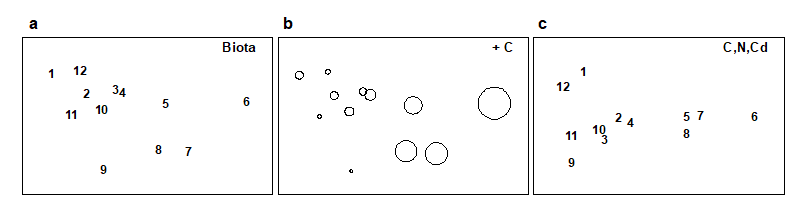
Methods which link multivariate biotic patterns to environmental variables are explored in Chapter 11; these are illustrated here by the Garroch Head dump-ground study described earlier (Fig. 1.5). The MDS of the macrofaunal communities from the 12 sites is shown in Fig. 1.11a; this is based on Bray-Curtis similarities computed from (transformed) species biomass values.ꞎ
Fig. 1.11. Garroch Head macrofauna {G}. a) MDS ordination of Bray-Curtis similarities from $\sqrt{}$-transformed species biomass data for the sites shown in Fig. 1.5; b) the same MDS but with superimposed circles of increasing size, representing increasing carbon concentrations in matched sediment samples; c) ordination of (log-transformed) carbon, nitrogen and cadmium concentrations in the sediments at the 12 sites (2-d MDS stress = 0.05).
Steady change in the community is apparent as the dump centre (site 6) is approached along the western arm of the transect (sites 1 to 6), then with a mirrored structure along the eastern arm (sites 6 to 12), so that the samples from the two ends of the transect have similar species composition. That this biotic pattern correlates with the organic loading of the sediments can best be seen by superimposing the values for a single environmental variable, such as Carbon concentration, on the MDS configuration. The bubble plot of Fig. 1.11b represents C values by circles of differing diameter, placed at the corresponding site locations on the MDS, and the pattern across sites of the 11 available environmental variables (sediment concentrations of C, N, Cu, Cd, Zn, Ni, etc.) can be viewed in this way (Chapter 11). This either uses a single abiotic variable at a time or displays several at once, as vectors – usually unsatisfactorily because it assumes a linear relationship of the variable to the biotic ordination points – or (more satisfactorily) by segmented bubble plots in which each variable is only a circle segment, of different sizes but at the same position on the circle (of the type seen in Figs. 7.14-16; see also Purcell, Rushworth, Clarke et al. (2014) .Ɥ
Where bubble plots are not adequate, because the 2- or 3-d MDS is a poor approximation (high stress) to the biotic similarity matrix, an alternative technique is that of linkage trees (multivariate regression trees), which carry out constrained binary divisive clustering on the biotic similarities, each division of the samples (into ever smaller groups) being permitted only where it has an ‘explanation’ in terms of an inequality on one of the abiotic variables (Chapter 11), e.g. “group A splits into B and C because all sites in group B have salinity > 20ppt but all in group C have salinity < 20ppt” and this gives the maximal separation of site A communities into two groups. Stopping the search for new divisions uses the SIMPROF tests that were mentioned earlier, in relation to unconstrained cluster methods (for a LINKTREE example see Fig. 11.14).
A different approach is required in order to answer questions about combinations of environmental variables, for example to what extent the biotic pattern can be ‘explained’ by knowledge of the full set, or a subset, of the abiotic variables. Though there is clearly one strong underlying gradient in Fig. 1.11a (horizontal axis), corresponding to an increasing level of organic enrichment, there are nonetheless secondary community differences (e.g. on the vertical axis) which may be amenable to explanation by metal concentration differences, for example. The heuristic approach adopted here is to display the multivariate pattern of the environmental data, ask to what extent it matches the between-site relationships observed in the biota, and then maximise some matching coefficient between the two, by examining possible subsets of the abiotic variables (the BEST procedure, Chapters 11 and 16).ȸ
Fig. 1.11c is based on this optimal subset for the Garroch Head sediment variables, namely (C, N, Cd). It is an MDS plot, using Euclidean distance for its dissimilarities,Ͷ and is seen to replicate the pattern in Fig. 1.11a rather closely. In fact, the optimal match is determined by correlating the underlying dissimilarity matrices rather than the ordinations themselves, in parallel with the reasoning behind the ANOSIM tests, seen earlier.
The suggestion is therefore that the biotic pattern of the Garroch Head sites is associated not just with an organic enrichment gradient but also with a particular heavy metal. It is important, however, to realise the limitations of such an ‘explanation’. Firstly, there are usually other combinations of abiotic variables which will correlate nearly as well with the biotic pattern, particularly as here when the environmental variables are strongly inter-correlated amongst themselves. Secondly, there can be no direct implication of causality of the link between these abiotic variables and the community structure, based solely on field survey data: the real driving factors could be unmeasured but happen to correlate highly with the variables identified as producing the optimal match. This is a general feature of inference from purely observational studies and can only be avoided formally by ‘randomising out’ effects of unmeasured variables; this requires random allocation of treatments to observational units for field or laboratory-based community experiments (Chapter 12).
† Though PRIMER offers nearly 50 of the (dis)similarity/distance measures that have been proposed in the literature.
¶ The PRIMER routines automatically offer this set of transformation choices, applied to the whole data matrix, but also cater for more selective transformations of particular sets of variables, as is often appropriate to environmental rather than species data.
§ Other ordination techniques in common use include: Principal Co-ordinates Analysis, PCO; Detrended Correspondence Analysis, DCA. Chapter 5 has some brief remarks on their relation to PCA and nMDS/mMDS but this manual concentrates on PCA and MDS, found in PRIMER; PCO is available in PERMANOVA+.
‡ PRIMER now performs tests for all 1-, 2- and 3-way crossed and/or nested combinations of factors in its ANOSIM routine, also including a more indirect test, with a different form of statistic, for factors (with sufficient levels) which do not have replication within their levels. These are all robust, non-parametric (rank-based) tests and therefore do not permit the (metric) partition of overall effects into ‘main’ and ‘interaction’ components. Within a semi-parametric framework (and still by permutation testing), such partitions are achieved by the PERMANOVA routine within the PERMANOVA+ add-on to PRIMER, Anderson, Gorley & Clarke (2008) .
⸸ This has been rectified in PRIMER 7, with its greater emphasis on species analyses, such as Shade plots, SIMPROF tests for coherent species groups, segmented bubble plots etc (Chapter 7).
⸙ SIMPER in PRIMER first tabulates species contributions to the average similarity of samples within each group then of average dissimilarity between all pairs of groups. Two-way and (squared) Euclidean distance options are given, the latter for abiotic data.
ꞎ Chapter 13, and the meta-analysis section in Chapter 15, discuss the relative merits and drawbacks of using species abundance or biomass when both are available; in fact, Chapter 13 is a wider discussion of the advantages of sampling particular components of the marine biota, for a study on the effects of pollutants.
Ɥ The PRIMER ‘bubble plot’ overlay can be on any ordination type, in 2- or 3-d, and has flexible colour/scaling options, as well as some scope for using a supplied image as the overlay.
ȸ The BEST/Bio-Env option in PRIMER optimises the match by examining all combinations of abiotic variables. Where this is not computationally feasible, the BEST/BVStep option performs a stepwise search, adding (or subtracting) single abiotic variables at each step, much as in stepwise multiple regression. Avoidance of a full search permits a generalisation to pattern-matching scenarios other than abiotic-to-biotic, e.g. BVStep can select a subset of species whose multivariate structure matches, to a high degree, the pattern for the full set of species (Chapter 16), thus indicating influential species or potential surrogates for the full community.
Ͷ It is, though, virtually indistinguishable in this case from a PCA, because of the small number of variables and the implicit use of the same dissimilarity matrix for both techniques.