Average body mass matrix (B/A)
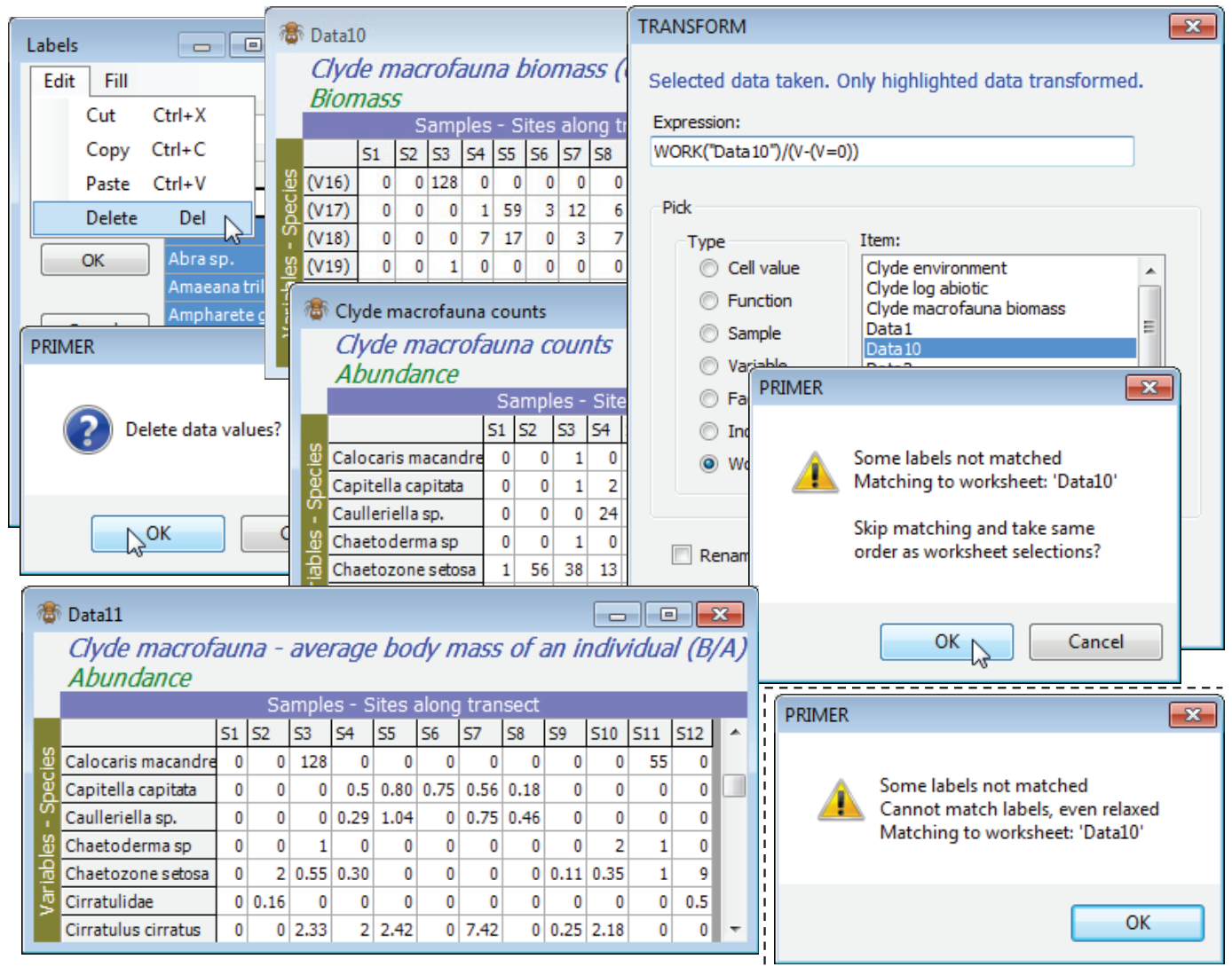
A useful variation of this, but one which needs more care, is to compute average body mass of each species in each sample. This is simply B/A, but needs to cater for the many cases when A (and B) are zero and a simple ratio is undefined. With active sheet Clyde macrofauna counts, so that V is again the counts, Pre-treatment>Transform(individual)>Expression: Work(“Clyde macrofauna biomass”)/(V – (V=0)) will do the trick, because when V>0 the expression (V=0) gives the value 0 (false), so that the correct ratio of B/A is calculated. However, when V=0 the expression (V=0) returns the value -1 (true). The bottom line is then 1 and the result of the ratio is a reasonable value of 0. This assumes that B=0 when A=0 of course! [This, incidentally, is something that can be checked by running Abundance-Biomass Comparison curves, described in Section 16, since the Analyse>Dominance Plot (ABC) routine explicitly checks for incorrect matrix entries which have A=0 but B>0; the converse is perfectly permissible – the weight of all organisms of a species in a sample might be too small to register – but this does not cause a problem with a B/A calculation).]
An illustration of error trapping and relaxation of strict matching, in Pre-treatment>Transform (individual) with matching of entries, is obtained by copying Clyde macrofauna biomass with Tools>Duplicate, then Edit>Labels>Variables on this to delete all the species labels (click the Label header and hit the delete key or Edit>Delete). A sheet cannot function without labels so PRIMER substitutes its own defaults of (V1), (V2), etc. Now run the above calculation on Clyde macrofauna counts, but with the relabelled biomass sheet (Data10 below) replacing the original biomass sheet. A warning message says that it could not find (variable) labels to match, but the two matrices are the same size so the option is given of proceeding anyway, on the assumption that the species order matches. We know it does here, so continue, to give the desired B/A matrix, and the original species labels will be present in the resulting new sheet because these are always taken from the active matrix, in a case such as this. Re-run having deselected one of the rows in Data10, however, and an irrecoverable error message occurs – a match is impossible because the variable labels do not match and neither does the number of variables in the A and B matrices.