Combined nMDS; (Messolongi diatoms & abiotic data)
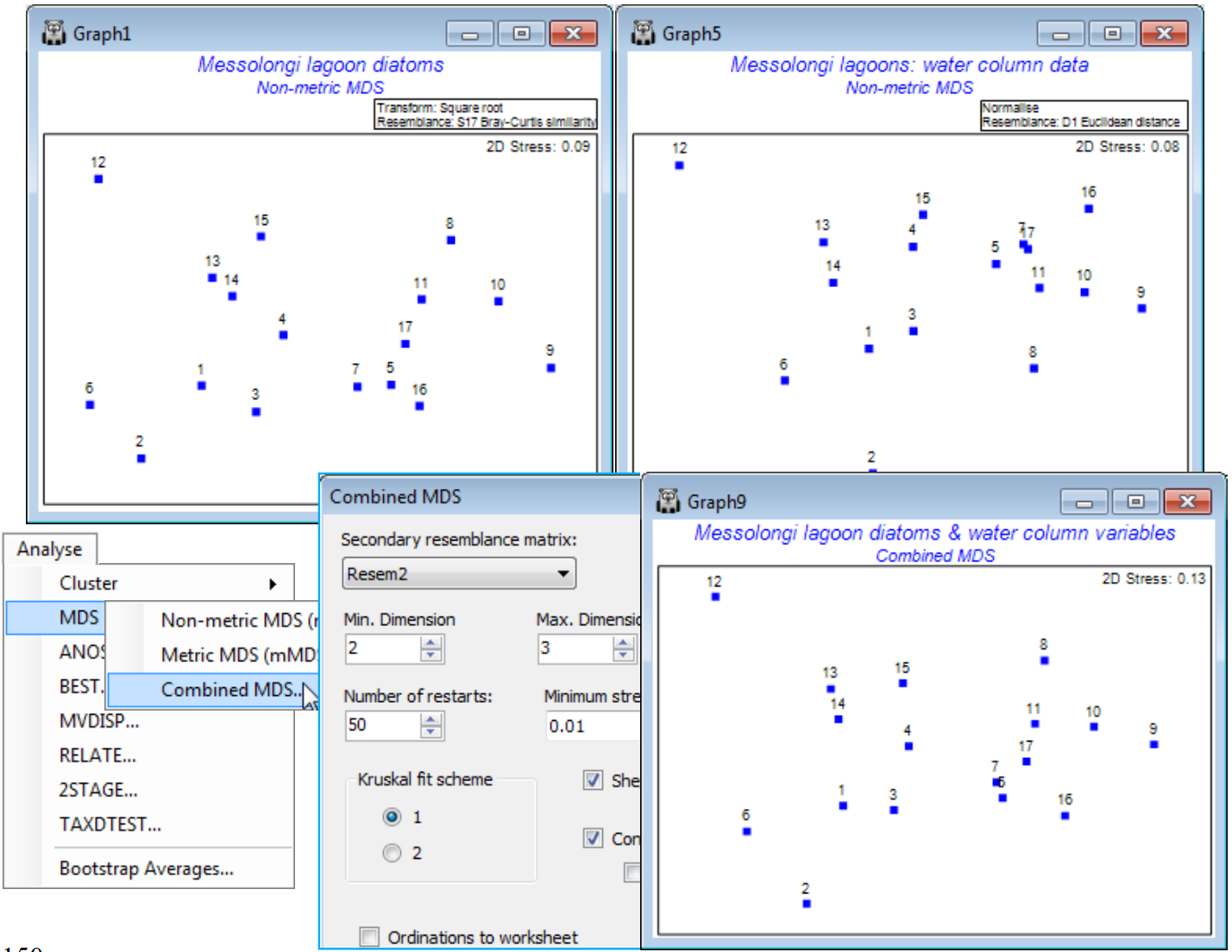
A variation of the above, and a final new PRIMER 7 feature to discuss in this section, is the option to mix, in equal measure, stress functions from two resemblance matrices, in a single nMDS. There are possible applications to data sets in which a single MDS is required from two different types of data, which may need separate dissimilarity definitions, e.g. perhaps in a ‘biotope’ ordination, data might consist of biotic communities, usually analysed with a Bray-Curtis similarity, and continuous environmental variables which would typically be normalised and input to Euclidean distance. The practical reality is that it would still be better to somehow build those into a common matrix, which could be analysed with the same resemblance measure, because then all the analyses based on a single triangular matrix (clustering, ANOSIM tests etc) would become available. But if the sole purpose is to produce an ordination which combines two resemblance matrices for the same set of sample labels (same sites, times etc) then Analyse>MDS>Combined MDS offers this possibility. The dialog is more or less the same as that for nMDS except for specification of the (Secondary resemblance matrix: ) to mix with that for the active sheet. A solution is sought, in the usual range of possible dimensions, by minimising an equal mix of the two stress functions, and the combined MDS is displayed along with the Shepard diagram for resemblances from the active sheet (to get the other Shepard diagram, re-run the routine with the ‘secondary’ resemblance matrix now as the active sheet – the stress value quoted on both plots will be the overall average stress). It is inevitable that the overall stress will be higher than that for the two analyses separately, because the routine is trying to compromise between a configuration of samples which would best suit each matrix separately; the stress only matches those of the individual solutions when they are identical.
A study of water column diatom assemblages (193 species) at 17 sites in the Messolongi lagoons of E Central Greece, and a matching set of 11 water quality variables measured at the same sites (with salinity fluctuating between brackish water and fully marine, and nutrient enrichment within certain lagoons) is looked at in detail in Section 13, on methods for biota-environment linkage. If analyses given there for the biotic data (root transform of diatom densities, followed by Bray-Curtis) and abiotic variables (log transform for all water column chemistry variables, excepting Temperature, Salinity, DO$_2 $ and pH, followed by normalisation and Euclidean distance) are repeated here, they lead to the separate nMDS plots shown below, with respective stress 0.09 and 0.08. There is a remarkable similarity in the patterns of the 17 sites between the two analyses, and running Analyse >MDS>Combined MDS on the Bray-Curtis similarity, supplying the Euclidean distance as the secondary resemblance matrix, leads to the combined nMDS shown, with stress of 0.13.