Check on aggregation files
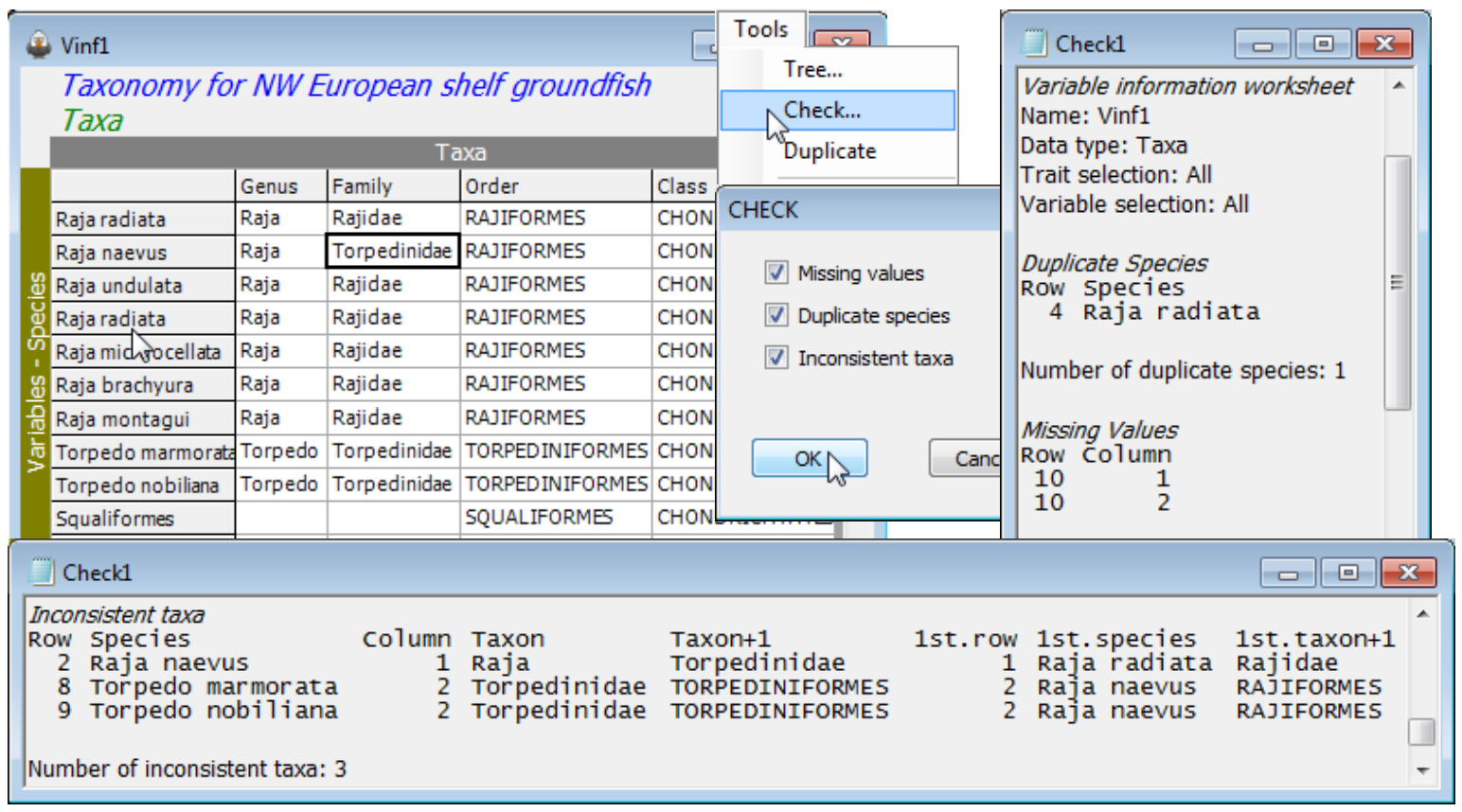
Use the open aggregation file, Groundfish taxonomy, to show the smaller set of Tools items (Tree, Check, Duplicate) available when the active window is of variable information. Tools>Duplicate has been seen previously for worksheets and plots (in Sections 3 and 8). Here it has the same effect, taking a copy of the Groundfish taxonomy window, called Vinf1, to the head of a fresh branch in the Explorer tree. Insert the following errors in Vinf1 to demonstrate the Tools>Check option:
a) overwrite Raja clavata (row 4) in the Species column with Raja radiata (by taking Edit>Labels>Variables and double clicking in the Raja clavata label and typing in the incorrect name);
b) whilst in the Labels dialog change Squalus acanthias (row 10) to Squaliformes (note that upper or lower case does not make a difference when matching names), and OK to exit back to the Vinf1 sheet, then delete Squalus and Squalidae from the genus and family name for that taxon;
c) change Rajidae to Torpedinidae as the family name for Raja naevus (row 2).
(Note that the row/column numbers of an entry can be found by clicking on it – the status bar at the bottom right displays the current cursor position). Then Tools>Check finds three types or error:
-
Duplicate Species in row 4 (the repeat of Raja radiata) – labels (samples or variables) should always be unique in a PRIMER worksheet, otherwise matching conflicts can easily result;
-
Missing Values (blanks) in row 10. This represents a common situation where only coarser-scale identifications can be made for some taxa. Nonetheless, aggregation sheets need to be complete, in order to avoid incorrect matching. E.g. another species from a completely different order but with a blank family (and genus) entry would be pooled with the Squaliformes abundance when the matrix is aggregated to genus or family level, because both entries have the same (blank) family name. Similar problems would occur with taxonomic distinctness calculations (Section 15). So blank entries should be filled with the names from the immediate right or left, depending on the context (often it make sense to fill from right to left). Here put Squaliformes in the two blanks – the routine does not object to the same name being used in different taxonomic levels.
-
Inconsistent taxa in rows 2, 8 and 9. In fact there is only one mistake, the family identification of Raja naevus, picked up in the correct row (2) because Raja has been established by row 1 to be a genus name in the Rajidae family, thus cannot also be a genus name in the family Torpedinidae. Quite often, however, an error is not discovered until a conflict occurs much later in the sheet, on a row which may be correct. This is seen in the inconsistent identification of the two Torpedo genera though neither are wrong. PRIMER 7 has greatly improved its diagnostics here, by listing not just the row and column on which the conflict occurred (in the first 5 columns of the output: row, species name, column, entry in that column, entry in the following column) but also what the conflict with an earlier row was (in the final three columns of the output: the earlier row, its species name, the entry in the earlier row causing the current conflict). So, Torpedo marmorata (row 8) in family Torpedinidae cannot be in order Torpediniformes because in a previous row (it identifies row 2) the family Torpedinidae were given as in order Rajiformes. With this level of diagnostics, errors in aggregation files (they commonly occur!) should be more easily fixed.