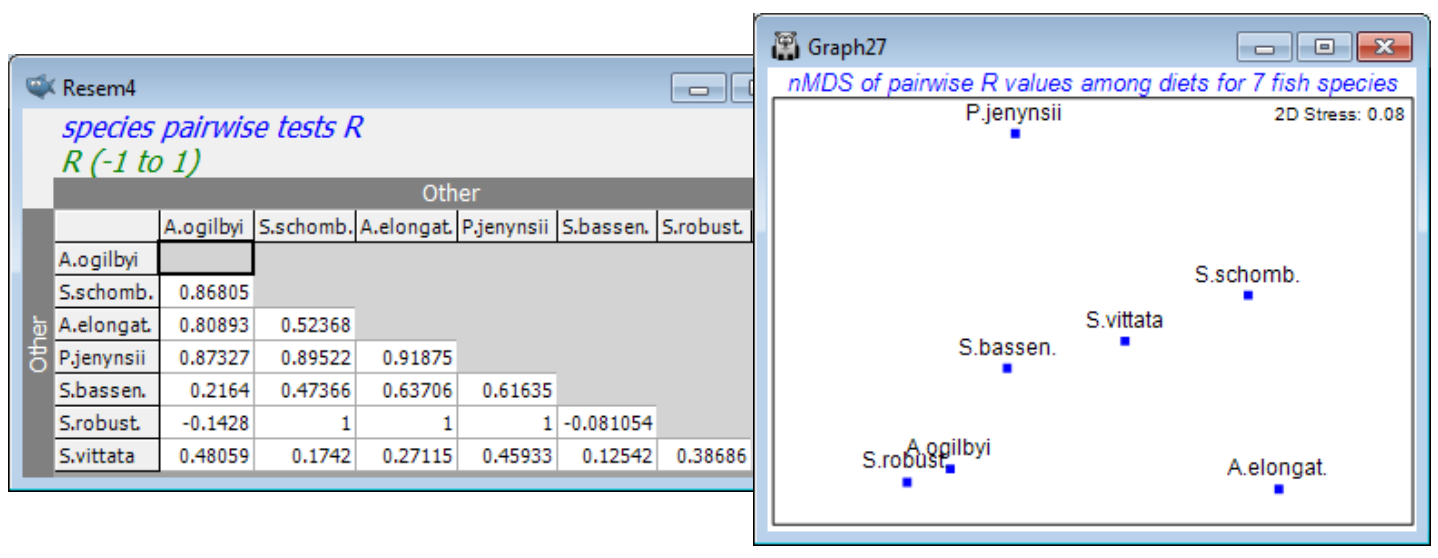
Other 1-way ANOSIM options
Checking the (✓Pairwise tests to worksheet) box has also sent the above R values to a worksheet in triangular format, which could be a useful layout for tabulating ANOSIM results in a publication. More subtly, this can be regarded as a resemblance matrix (of distance-type) in its own right – the higher the value of R the greater the separation of replicates from two groups in the high-d (prey) space. Inputting this to an MDS plot will display the relationships between these 7 groups, and can be seen as a type of means plot. [Note that this triangular array is not a sensible distance matrix at present because it can, and does, contain (small) negative values. Input to metric MDS without some prior rescaling would be problematic therefore. However, nMDS effectively works only on the rank orders of the entries so there is no need to rescale them – the lowest values (the negative ones) indicate the least established differences in diet and the highest values (R=1) the greatest differences, which is exactly what is required for a sensible nMDS plot here. Dropping the negative signs, by taking absolute values of the entries, would not be the technically correct approach here.]
The more straightforward means plot is, as we have seen before, is to average the replicates, and then calculate Bray-Curtis between these mean dietary samples, ordinating by nMDS or mMDS. But there are many other possibilities for a direct means plot! The data could be transformed before or after averaging, or the dissimilarities could be averaged – or even their ranks averaged. PRIMER 7 now has the option to average (dis)similarities across a group structure, with Tools>Average for an active window of a resemblance matrix. Tools>Rank distance will also replace resemblance entries with their ranks. (A further option is given in the PERMANOVA+ add-on, of computing distances among centroids in the high-d PCO space formed from the resemblance matrix). These will all give means plots with slightly different emphases. In the case of the matrix of R values, this highlights relative group separations, i.e. adjusting differences by within-group dispersion.
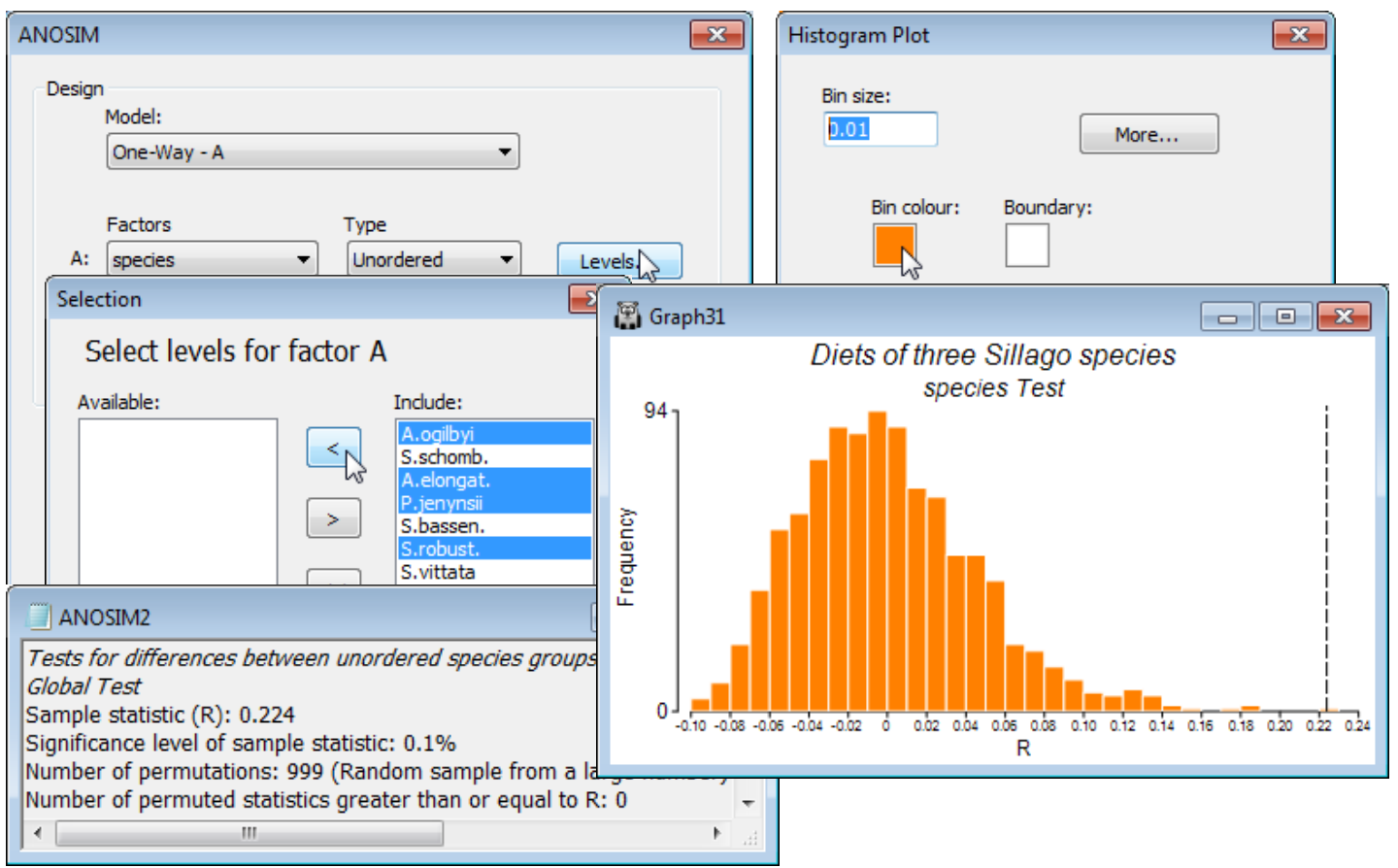
Other options within the ANOSIM routine include the ability to manipulate the histogram for the global R statistic by rescaling axes, titles etc (the usual Graph>Sample Labels & Symbols menu) and changing bin widths and, in v7, bin colours (Graph>Special), as for any other histogram plot. There is also a check box in the ANOSIM dialog to send (✓R values to file). You would then need to supply an *.txt file name which will hold a simple list – one number to a line, in simple text – of the R values for the 999 (or however many) permutations carried out for the global test. This would allow the null distribution data to be replotted, for example, in another statistical/graphical package.
As noted earlier, the plotted histogram (and the listed R values) refer only to the global test for no differences among any of the groups. If you require a histogram for a specific pairwise comparison then you will need to pick out that pair of groups and re-run ANOSIM, selecting either externally, by Select>Samples on the original resemblance matrix, or internally, using the Levels button for A on the ANOSIM dialog. Both lead to the usual Selection dialog. For a pairwise test, it will make no difference to the R value (or to its significance level) whether the results are read from the above pairwise table or recalculated with just those groups selected, so this would only be useful: a) if you required the pairwise histogram, or b) a test for a specific subset of three groups, four groups etc was needed. As seen in Section 3, a relevant a priori hypothesis here concerns whether there are detectable dietary differences between the three congeneric Sillago fish species (S. schomburgkii, S. bassensis and S. vittata). After testing this, save and close the workspace WA fish ws.