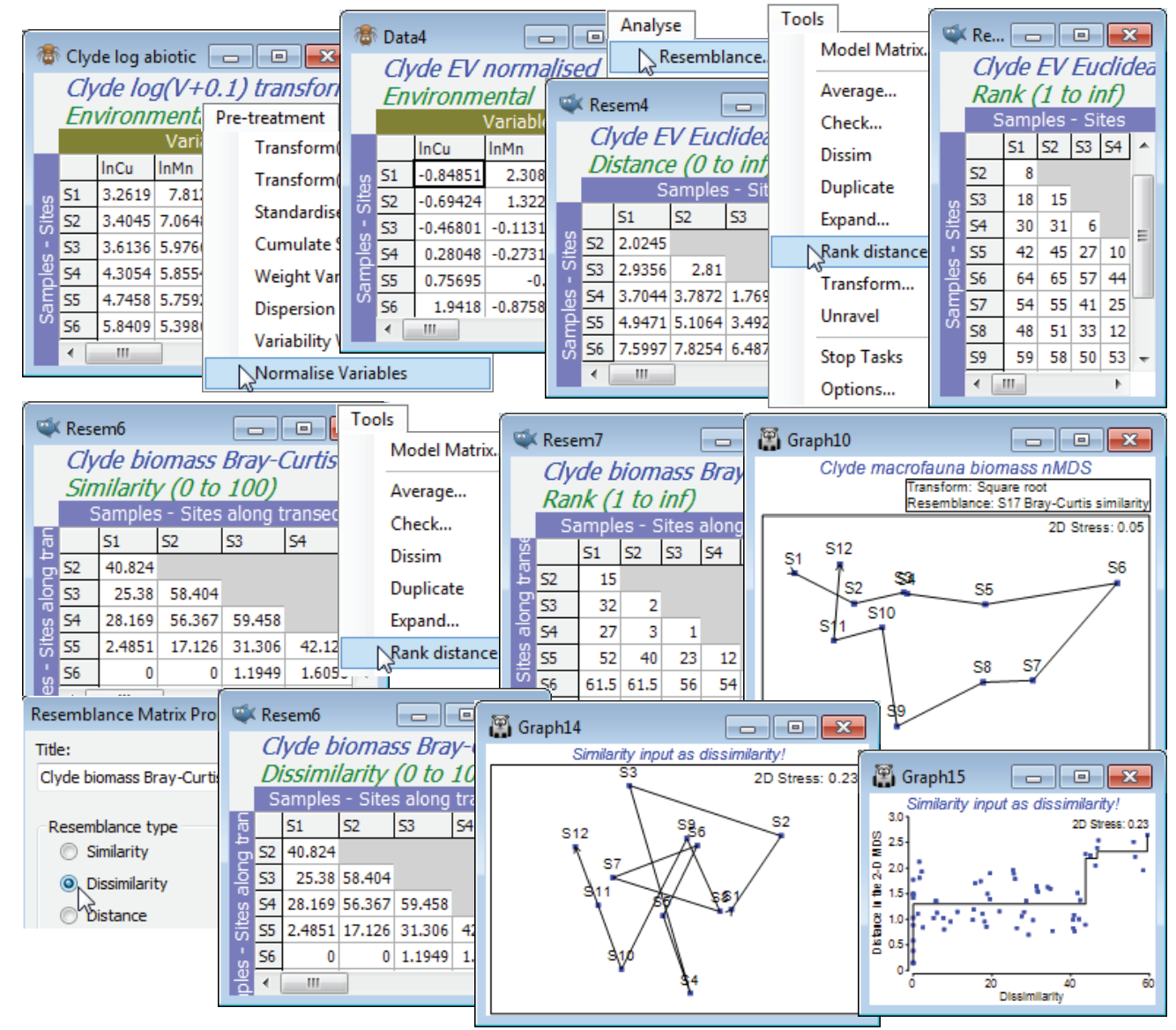
Ranked resemblances
Ranking is also a menu option when the active sheet is a resemblance (Tools>Rank distance), but it operates a little differently. This time, all elements of the triangular matrix are ranked together, rather than separate ranking of the rows or columns of the rectangular data sheet. Do not get these two possible rankings confused! It is easy to fall into the trap of thinking that, because a ranked data matrix will be the same whether ranked from original or transformed data, if you are intending to rank the similarity matrix then initial transformation of the data does not matter. This is entirely wrong of course – ranking the similarity matrix is by no means the same as ranking the data then calculating the similarities! In fact, whilst ranking the data may play a marginally useful role for handling outliers in environmental matrices (as above), it rarely makes sense for assemblage data because it destroys the special nature of the (very many) zero responses, which would be assigned different tied ranks for different species. Ranking the resemblances, however, is rather central to the approach in PRIMER: many of the core routines (ANOSIM, RELATE, BEST, …) start from the ranked form of the similarity matrix, and nMDS ordination also exploits this rank order. For all routines, however, it is not necessary to enter the ranked form of the triangular matrix – if the result depends only on the ranks, this will be part of the internal calculation on the similarities. The menu item of Tools>Rank distance on a resemblance matrix is mainly here to help visualise and check the relatively simple computations underlying an ANOSIM test, for example (see the definition of the ANOSIM R statistic, a difference in mean rank dissimilarities, in equation 6.1 of CiMC).
For the Euclidean distance matrix from the above Clyde environmental data (transformed then normalised), take Tools>Rank distance to produce a rank resemblance matrix. Note that entries are just the numbers 1, 2, .., 66. Importantly, the convention PRIMER adopts here is always to return a distance-type matrix from the Rank distance operation, irrespective of whether it is given a similarity or dissimilarity/distance matrix (explaining why the menu item is called Tools>Rank distance). Thus rank 1 corresponds to samples (S11, S12), which are closest environmentally, and rank 66 to those furthest apart (S6, S9). To see this point about the direction in which ranks are assigned, open the macrofaunal biomass matrix for the 12 samples on the Clyde transect, Clyde macrofauna biomass, take a square transform and calculate Bray-Curtis similarity, as usual. Now take Tools>Rank distance on this similarity matrix and note that the resulting ranks again form a distance matrix, with the closest sites in assemblage terms (rank 1) being S3 and S4, and several pairs of sites tied on the largest, most distant rank (average of 61.5), namely S6 with S1, S2, S11, S12 etc, which are all pairs of sites with no species in common.
PRIMER handles its (distance) ranks in this slightly unconventional way to reassure the user that, on the many occasions when two sets of resemblances are compared to see if they are arranging the samples in a similar high-dimensional pattern (e.g. assemblage vs environment, Bray-Curtis vs Chi-squared or Euclidean measures, biomarkers vs tissue burdens etc), the user does not have to worry whether the two resemblance matrices are the ‘same way round’ (whether high values correspond to large or small differences between samples). This is always adjusted to the correct comparison, in the same way that the MDS routine will always internally turn a similarity into a dissimilarity when it is matching this up to distance in the ordination space (as in the Shepard diagram). You can force PRIMER to do the stupid thing, e.g. run MDS the ‘wrong way round’, making it try to place sites that should be similar at the greatest distance apart and sites that have little in common close together (with resultant very high stress levels, and a crazy plot and Shepard diagram!). But you can only do this by giving PRIMER a genuine similarity matrix and calling it a dissimilarity, by using Edit>Properties to change (Resemblance type•Similarity) to (•Dissimilarity).