(Fal estuary copepods)
Sediment copepod assemblages (and other fauna) from five creeks of the Fal estuary, SW England, were analysed by Somerfield PJ, Gee JM, Warwick RM 1994, Mar Ecol Prog Ser 105: 79-88. The sediments of this estuary are characterised by high and varying concentrations of heavy metals, a result of tin and copper mining over hundreds of years. The copepod data consist of 23 species found in 27 samples, consisting of 5 replicate cores spanning each creek (Mylor: M1-M5; Pill: P1-P5; St Just: J1-J5; Percuil: E1-E5; and 7 from the largest creek, Restronguet: R1-R7). These are in directory C:\Examples v7\Fal benthic fauna, worksheet Fal copepod counts(.pri), with a factor Creek identifying samples from the 5 creeks. There are also environmental cores (of silt/clay ratios, heavy metals etc) matching these 27 sample locations, held in an Excel file Fal environment(.xls), plus nematode densities, macrofaunal counts and biomass, and associated aggregation files.
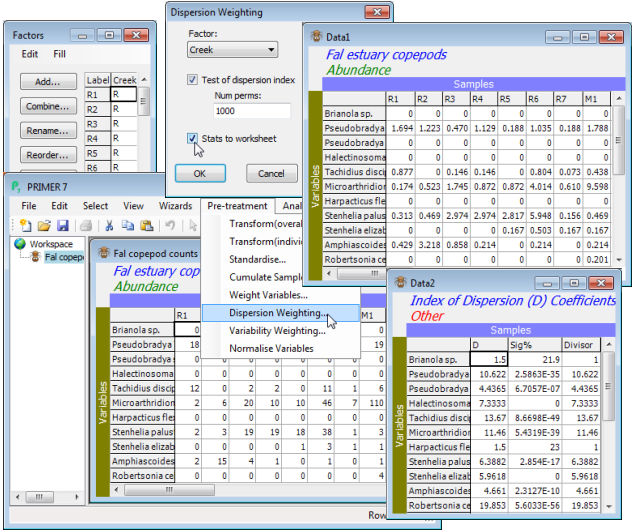
File>Open the copepod data and take Pre-treatment>Dispersion weighting>(Factor: Creek) & (✓Test of dispersion index) & (Num perms: 1000) & (✓Stats to worksheet). The Data1 sheet gives the dispersion weighted counts, which are either ready to go into the Analyse>Resemblance step of the next section, or could be mildly transformed before they do so, as shown earlier with Pre-treatment>Transform(overall)>(Transformation: Square root). There seems little need for the latter, however, since the dispersion weighting has already succeeded in downweighting the larger, erratic counts coming from P. littoralis, R. celtica, E. gariene and T. discipes and the somewhat less erratic P. curticorne and M. falla – the matrix Data1 now has no dispersion-weighted ‘counts’ in double figures, and the subsequent untransformed analysis will not be dominated by a small set of species. In three columns, Data2 gives: the mean dispersion indices $\overline{D}$ for each species; the evidence for clumping (i.e. the % significance level for a test of $\overline{D} = 1$); and the actual divisor used for that species row, which is 1 if the test does not reject this hypothesis at 5% (or better). Thus, T. discipes values are divided by 13.67 but Brianola sp. remains unchanged, though $\overline{D} = 1.5$. You might now like to run the routine again for the Fal nematode abundance file, which inspection shows must be numbers scaled up to a density, not real counts (e.g. there are no entries of 1!). The tick box for the test must be unchecked, the resulting $\overline{D}$ values are all >>1, but weighting by $\overline{D}$ is still justifiable.