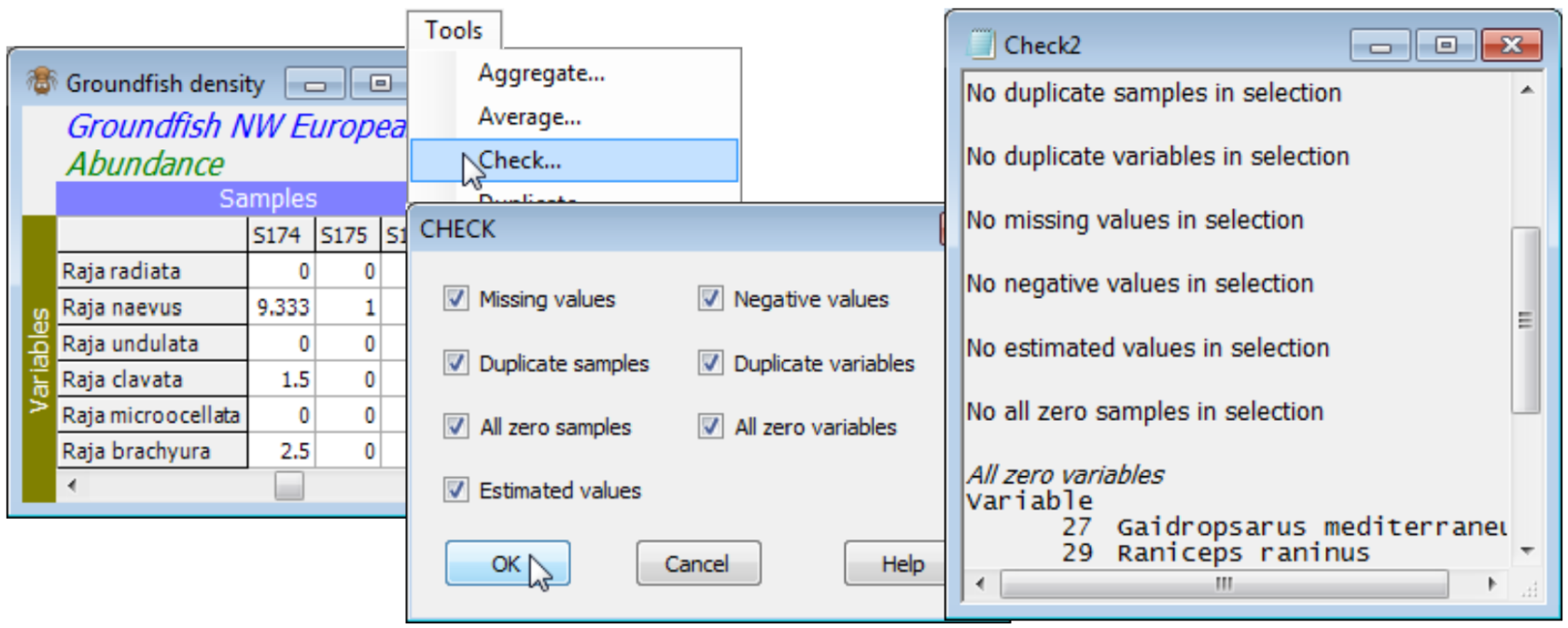
Tree menu; Check on datasheets & resemblances; Undefined resemblances
When the active window is a datasheet, Tools>Check can check for the following: a) ✓Missing values, identified in the sheet by ‘Missing!’, and which might have been read in as blank cells in an Excel worksheet for example; b) ✓Negative values, which are not appropriate for abundance-type data analysed by Bray-Curtis, though common for environmental variables (especially normalised) input to Euclidean distance; c) ✓Duplicate sample (and/or) variable labels, which are tolerated for some analyses (warnings are usually given) but are best avoided wherever possible; d) ✓All zero samples (and/or) variables; and e) ✓Estimated values, displayed in red type in the matrix. The latter come from applying Tools>Missing (seen shortly) to environmental variables – or to other normally distributed data – containing Missing! cells, which otherwise might not be tolerated by some analysis routines requiring complete data. All or any of the 7 boxes can be ticked. Whether it is important to check for a particular attribute depends on the analysis. For example, species which are zero over all samples will be ignored when Bray-Curtis similarity is computed among samples, and can safely be left in the matrix, but all-zero samples are potentially more of a problem since Bray-Curtis similarity between two blank samples is set to ‘Undefined!’. Dependent on the context, these samples might best be omitted, or a different similarity used (e.g. zero-adjusted Bray-Curtis, Section 5), or the entry left as ‘Undefined!’, i.e. treated as unknown.
When the active window is a resemblance sheet, Tools>Check looks for only three data attributes: a) ✓Undefined values, arising as suggested above; b) ✓Out of bounds values, for distance coeff-icients (or transformations) that return very large or small values (NaN); and c) ✓Duplicate labels, as above. Blanking a cell in a resemblance matrix sets it to Undefined! status, and several of the core routines using resemblances (e.g. MDS, Cluster, ANOSIM) are carefully written in PRIMER to tolerate a few Undefined! entries, treating them as unknown. (You can appreciate that knowing the similarities $S_{12}$, $S_{13}$, $S_{14}$, $S_{23}$, $S_{24}$ might enable you to place four samples in relation to each other without knowing similarity $S_{34}$). Blanking out NaN (Not a Number) entries, to Undefined!, is one possibility therefore, but others may be equally good or better (replacing by a large, but finite value, modifying the coefficient or transformation which generated them etc).